Abstract
Anaplasma marginale (order Rickettsiales, family Anaplasmataceae), a tick-borne pathogen of cattle, is endemic in tropical and subtropical regions of the world. Many geographic isolates of A. marginale occur in the United States and have been identified by major surface protein 1a (MSP1a), which varies in sequence and molecular weight due to different numbers of tandem 28- to 29-amino-acid repeats. The present study was undertaken to examine the genetic variations among isolates of A. marginale obtained during 2001 from infected cattle from east-central Oklahoma, where A. marginale is endemic. The gene and protein sequences of MSP1a and msp4 nucleotide sequences were used to infer the phylogenetic relationships among Oklahoma and New World isolates from Argentina, Brazil, Mexico, and the United States. All 11 A. marginale isolates collected from Oklahoma had different MSP1a sequences but identical MSP4 sequences. The phylogenies of the msp4 sequences of 13 isolates from Oklahoma in comparison with those of 7 Latin American isolates and 12 U.S. isolates by maximum-parsimony (MP) and maximum-likelihood (ML) analyses, with A. centrale and A. ovis sequences used as outgroups, provided strong bootstrap analysis support for a Latin American clade. Isolates of A. marginale from the southern United States (Florida, Mississippi, and Virginia) and the west-central United States (California, Idaho, Illinois, Oregon, Missouri, and Texas) also grouped into two clades. Both clades contained isolates from Oklahoma, suggesting extensive cattle movement. ML analysis of the msp4 sequences of isolates from Oklahoma provided bootstrap analysis support for east-central and north-central clades in Oklahoma, and both clades included isolates from Stillwater, Okla. Analysis of the codon and amino acid changes among the msp4 sequences of isolates with different phylogenies provided evidence that msp4 is not under positive selection pressure. In contrast, the phylogenies of the MSP1a DNA and protein sequences of 13 isolates from Oklahoma in comparison with those of 7 Latin American and 13 isolates from the United States by MP and ML analyses demonstrated no geographic clustering and provided evidence that this gene is under positive selection pressure. The results indicate that msp1α is not a marker for the characterization of A. marginale geographic isolates and suggest that the genetic heterogeneity observed among isolates of A. marginale within Oklahoma could be explained by cattle movement and the maintenance of different genotypes by independent transmission events.
Anaplasma marginale is a rickettsial pathogen that causes the disease anaplasmosis in cattle (25). Feeding ticks effect biological transmission of this obligate intraerythrocytic organism, while mechanical transmission occurs when infected blood is transferred to susceptible cattle by biting flies or blood-contaminated fomites (for a review, see the report by Ewing [13]). Many A. marginale geographic isolates which differ in biology, morphology, protein sequence, and antigenic characteristics have been identified and have been characterized by the major surface protein 1a (MSP1a), which varies in sequence and molecular weight due to different numbers of tandem 28- to 29-amino-acid repeats (for a review, see reference 5). The MSP1a tandem repeats are located after a conserved decapeptide in the amino-terminal region of the protein and are exposed extracellularly for interaction with host cell receptors (11). The frequency of variable amino acid positions in geographic isolates is higher in this region than in the rest of the protein (for a review, see reference 5).
The organisms in the order Rickettsiales have been reclassified into two families, the Anaplasmataceae and Rickettsiaceae, on the basis of genetic analyses of the 16S rRNA, groesl, and surface protein genes (12). Recent research has focused on MSPs that are involved in interactions with vertebrate and invertebrate host cells (7, 8, 20, 21) and have been used to elucidate the phylogeographic patterns of A. marginale (5, 6, 9, 16, 19). These MSPs are involved in host-pathogen interactions and may evolve more rapidly than other nuclear genes because of selective pressures exerted by host immune systems. Of the six A. marginale MSPs that have been identified and characterized, only three (MSP1a, MSP4, and MSP5) are encoded by single genes. Because these MSPs do not appear to undergo antigenic variation in cattle or ticks (4), they were posited to be more stable genes for phylogenetic studies. MSP1a, encoded by msp1α, has been reported to be an adhesin for bovine erythrocytes and tick cells and to effect adhesion, infection, and transmission of A. marginale by ticks of the genus Dermacentor (7, 8, 20, 21). Although the specific function of MSP4 is not known, our previous analysis of the msp4 gene from A. marginale isolates demonstrated that its sequence varies sufficiently among isolates to support its use in phylogeographic studies (6, 9).
Preliminary analysis of the phylogenetic relationships of A. marginale isolates on the basis of the MSP1a and MSP4 sequences demonstrated phylogeographic partitioning of A. marginale isolates in the United States (6). This information, together with the fact that for many isolates infection with geographic isolates is not cross protective, supports the inclusion of several isolates of A. marginale in the development of vaccine formulations for the United States (17). However, recent data support the existence of genetic heterogeneity in the structures of the msp1α sequences of isolates recovered from infected animals within a state (9) and even in a single herd of cattle in an area of endemicity (23), thus questioning the use of msp1α sequences to identify geographic isolates of A. marginale. Although single msp1α genotypes have been identified in individual cattle that were naturally or experimentally infected and sampled at different stages of infection (4, 23), a finding that we demonstrated could be explained by infection exclusion of A. marginale isolates in infected cattle (10), these findings suggest a rapid evolution of msp1α sequences, resulting in the maintenance of A. marginale genotypes in the population through different transmission events (9, 10, 23).
To test these hypotheses, we characterized the msp1α and msp4 genotypes of A. marginale isolates obtained during 2001 from infected cattle in east-central Oklahoma, where A. marginale is endemic, and conducted phylogenetic studies with two other isolates previously obtained from Oklahoma and isolates from Latin America and the United States.
MATERIALS AND METHODS
A. marginale isolates.
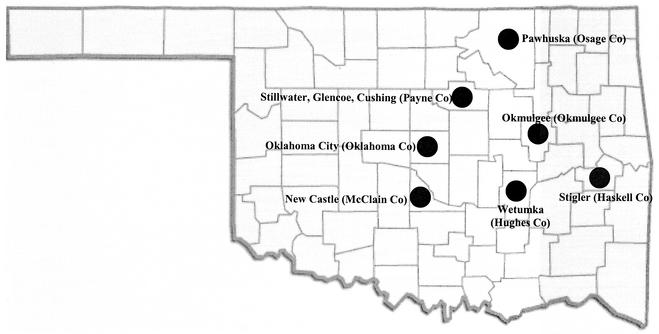
The A. marginale isolates obtained during 2001 from naturally infected cattle in Oklahoma plus the two previously characterized isolates from Wetumka and Pawhuska, Okla., used in this study are listed in Table 1. The geographic distributions of the isolates in Oklahoma are shown in Fig. 1. The Latin American isolates (from Mexico, Argentina, and Brazil) and the U.S. isolates of A. marginale, as well as the A. centrale and A. ovis isolates, were described previously (9); however, the isolate from Missouri was obtained from an infected cow in Buffalo, Mo., in 2001. The msp4 sequences of A. centrale (GenBank accession no. AF428090) and A. ovis (GenBank accession no. AF393742) and the msp1α sequence of an A. marginale isolate from Israel (GenBank accession no. AF352560) were used for outgroup comparisons.
TABLE 1.
Amplified sequences of A. marginale isolates from Oklahoma and their characteristicsa
| Isolate | Characteristics (location, host clinical parameters, date) | Genes (no. of repeats for MSP1a) | GenBank accession no. |
|---|---|---|---|
| Oklahoma | Wetumka, Hughes Co., 1997 | msp1α (3) | AY010247 |
| MSP4 | AY010252 | ||
| Glencoe 1 | Hesser Dairy; Holstein-Friesian herd; cow 23; Glencoe, Payne Co.; PCV = 15%; infection = 2.4%; Aug. 2001 | msp1α (5) | AY127053 |
| msp4 | AY127067 | ||
| Glencoe 2 | Hesser Dairy; Holstein-Friesian herd; cow 14; Glencoe, Payne Co.; PCV = 15%; infection = 1.4%; Sept. 2001 | msp1α (4) | AY127054 |
| msp4 | AY127068 | ||
| Glencoe 3 | Hesser Dairy; Holstein-Friesian herd; cow 242; Glencoe, Payne Co.; PCV = 18.5%; infection = 4.9%; Nov. 2001 | msp1α (3) | AY127055 |
| msp4 | AY127069 | ||
| Cushing 1 | Kuykendall Dairy; Aberdeen Angus herd; Cow “Dusty”; Cushing, Payne Co. PCV = 12.7%; infection = 59.9%; Sept. 2001 | msp1α (4) | AY127056 |
| msp4 | AY127070 | ||
| Cushing 2 | Kuykendall Dairy; Aberdeen Angus herd; cow “Rassy”; Cushing, Payne Co.; PCV = 35%; infection = 0.1%; Nov. 2001 | msp1α (5) | AY127057 |
| msp4 | AY127071 | ||
| Stigler | Jones Dairy; Stigler, Haskell Co; PCV = 10%; infection = 48%; Oct. 2001 | msp1α (4) | AY127058 |
| msp4 | AY127072 | ||
| Oklahoma City | Oklahoma City Market (now in Newalla, Oklahoma Co); PCV = 8.5%, infection = 9.8%; Oct. 2001 | msp1α (1) | AY127059 |
| msp4 | AY127073 | ||
| Okmulgee | Okmulgee, Okmulgee Co.; PCV = 11.1%, infection = 49.5%; Oct. 2001 | msp1α (4) | AY127060 |
| msp4 | AY127074 | ||
| Stillwater 1 | Myron dairy; cow 1; 823 N. Bethel, Stillwater, Payne Co.; PCV = 30%; infection = 0.3%; Aug. 2001 | msp1α (5) | AY127061 |
| msp4 | AY127075 | ||
| Stillwater 2 | Myron dairy; cow 2; 823 N. Bethel, Stillwater, Payne Co.; PCV = 18.2%; infection = 3.7%; Oct. 2001 | msp1α (4) | AY127062 |
| msp4 | AY127076 | ||
| New Castle | New Castle, McClain Co.; PCV = 10.5%; infection = 6.3%; Oct. 2001 | msp1α (4) | AY127063 |
| msp4 | AY127077 | ||
| Pawhuska | Pawhuska, Osage Co., 1960s | msp1α (2) | AY127064 |
| msp4 | AY127078 |
All isolates were obtained from naturally infected cattle. Isolates Oklahoma and Pawhuska were originally obtained from naturally infected cattle and for this study were isolated from bovine erythrocytes collected from experimentally infected cattle. Whole blood was collected in VACUTAINER tubes containing EDTA and used for preparation of stained blood smears for counting of percent infected erythrocytes (infection) by light microscopy and for determination of the packed cell volume (PCV).
FIG. 1.
Distribution by county of Oklahoma isolates of A. marginale used in the study.
A. marginale msp1α and msp4 PCR and sequencing.
A. marginale DNA was extracted from 0.5 to 1 ml of erythrocytes prepared from blood collected in EDTA-treated tubes as reported previously (6). The msp1α gene was amplified from 1 μl (1 to 10 ng) of DNA by PCR with 10 pmol of each primer (primer MSP1αP [5′-GCATTACAACGCAACGCTTGAG-3′] and primer MSP1α3 [5′-GCTTTACGCCGCCGCCTGCGCC-3′]) in a 50-μl volume (1.5 mM MgSO4, 0.2 mM deoxynucleoside triphosphates, 1× avian myeloblastosis virus-Tfl reaction buffer, 5 U of Tfl DNA polymerase) with the Access RT-PCR system (Promega, Madison, Wis.). The reactions were performed in an automated DNA thermal cycler (Mastercycler personal; Eppendorf, Westbury, N.Y.) for 35 cycles. After an initial denaturation step at 94°C for 30 s, each cycle consisted of denaturation at 94°C for 30 s and annealing-extension at 68°C for 2.5 min. The program ended by storage of the reaction mixtures at 4°C. The msp4 gene was amplified as described above but with oligonucleotides MSP45 (5′-GGGAGCTCCTATGAATTACAGAGAATTGTTTAC-3′) and MSP43 (5′-CCGGATCCTTAGCTGAACAGGAATCTTGC-3′) and a PCR profile of denaturation at 94°C for 30 s, annealing at 60°C for 30 s, and extension at 68°C for 1 min. The PCR products were electrophoresed on 1% agarose gels to check the sizes of the amplified fragments.
The amplified fragments were purified with resin (Wizard; Promega) and were cloned into the vector pGEM-T (Promega) or used directly for sequencing of both strands by double-stranded dye-termination cycle sequencing (Core Sequencing Facility, Department of Biochemistry and Molecular Biology, Noble Research Center, Oklahoma State University). The msp4 gene was completely sequenced. For the msp1α gene, only the fragment containing the upstream and variable regions was sequenced for all isolates (6, 9). When clones were recovered, at least two clones from each PCR were sequenced.
Sequence alignment and phylogenetic analysis.
The A. marginale msp1α gene fragment and the derived protein sequence containing the upstream and variable regions and the entire msp4-coding region were used for sequence alignment and phylogenetic analysis. Multiple-sequence alignment was performed with the program AlignX (Vector NTI Suite, version 5.5; InforMax, North Bethesda, Md.) with an engine based on the Clustal W algorithm (28). Nucleotides were coded as unordered, discrete characters with five possible character states (A, C, G, T, or N), and gaps were coded as missing data. Maximum-parsimony (MP) analyses were conducted by using equal weights for all characters and substitutions, heuristic searches with 25 random additions of input taxa, and tree bisection-reconnection (TBR) branch swapping by using the PAUP*4.0b4a program (27). The stability or the accuracy of the inferred topology(ies) was assessed by bootstrap analysis (14) of 200 iterations of heuristic searches with 25 random additions of input taxa and TBR branch swapping. Character-state changes were polarized by designating A. centrale and A. ovis as outgroups for the msp4 data set and by designating an A. marginale isolate from Israel as the outgroup for the msp1α and MSP1α data sets. To examine the effect of the analysis method on the resulting phylogenetic tree, we constructed a neighbor-joining phylogenetic tree (26) based on the distances obtained by maximum-likelihood (ML) analysis, with the model of DNA substitution chosen with MODELTEST (24).
To examine the phylogeographic patterns among the Oklahoma isolates of A. marginale, unrooted phylogenetic trees were constructed by ML analysis with the appropriate model of DNA sequence evolution, as determined with MODELTEST (24). The stabilities of the clades on the resulting msp4 and msp1α or MSP1a phylogenetic trees were estimated via 200 bootstrap iterations.
Selection pressure on msp1α and msp4.
The relative frequencies of nonsynonymous substitutions (dNs) and synonymous substitutions (dSs) among all pairwise comparisons of msp1α and msp4 sequences were estimated with MEGA (Molecular Evolutionary Genetics Analysis, version 1.01) software (18) by the method of Nei and Gojobori (22) and by application of the correction for multiple substitutions of Jukes and Cantor (15).
Nucleotide sequence accession numbers.
The GenBank accession numbers for the msp1α sequences of the Missouri and Oklahoma isolates of A. marginale are AY127052 to AY127064; and those for the msp4 sequences of the Oregon, Missouri, and Oklahoma A. marginale isolates are AY127065 to AY127078.
RESULTS
We sequenced the msp1α genes (512 to 861 bp; mean, 766 bp) and msp4 genes (851 bp) of 11 isolates of A. marginale collected from cattle in east-central Oklahoma during 2001. For comparative purposes, we included the DNA sequences of these two genes from two isolates of A. marginale previously collected in Wetumka, Okla., in 1997 (Oklahoma isolate) (5-10) and Pawhuska, Okla., in the 1960s (Table 1). All A. marginale isolates from Oklahoma possessed different msp1α and MSP1a sequences, with the number of tandem repeats varying from 1 (isolate Oklahoma City) to 5 (isolates Glencoe 1, Cushing 2, and Stillwater 1) (Fig. 2 and Table 1). Isolates of A. marginale from Glencoe and New Castle and one of the two isolates from Stillwater possessed identical msp4 sequences. Moreover, both isolates from Cushing possessed the same msp4 sequence as the isolate from Pawhuska that was collected in the 1960s. The remaining five isolates possessed unique msp4 sequences. All Oklahoma isolates of A. marginale had identical MSP4 sequences.
FIG. 2.
Comparison of the deduced amino acid sequences of the MSP1a variable region of Oklahoma isolates of A. marginale. The amino-terminal decapeptide and the beginning of the conserved region after the tandem repeats are underlined. The one-letter amino acid code is used. Asterisks indicate identical amino acids. Gaps indicate deletions or insertions.
To evaluate the phylogenetic affinities of the msp1α and msp4 sequences of the Oklahoma A. marginale isolates, we included sequences from 7 Latin American isolates and 13 (for msp1α) or 11 (for msp4) U.S. isolates of A. marginale in subsequent analyses. The mean number of differences in the msp1α sequences for all pairwise comparisons of A. marginale isolates from Oklahoma was 14.5 (Table 2), whereas the mean number of differences in the msp1α sequences for all pairwise comparisons of the New World isolates examined was 19.7. In contrast, only seven unique msp4 sequences were detected among the isolates from Oklahoma, with the mean number of DNA substitutions for all pairwise comparisons being 3.8 (Table 2), which is only slightly less than the mean number of differences for all pairwise comparisons of New World isolates examined (n = 4.6).
TABLE 2.
Absolute number of DNA sequence differences of msp1α and msp4 for all pairwise comparisons of Oklahoma A. marginale isolates
| Isolate | No. of DNA sequence differencesa
|
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Cushing 1 | Cushing 2 | Glencoe 1 | Glencoe 2 | Glencoe 3 | New Castle | Okla- homa | Oklahoma City | Okmulgee | Pawhuska | Stillwater 1 | Stillwater 2 | Stigler | |
| Cushing 1 | 25 | 24 | 19 | 7 | 5 | 19 | 11 | 12 | 21 | 25 | 3 | 9 | |
| Cushing 2 | 0 | 3 | 8 | 19 | 24 | 7 | 16 | 20 | 10 | 2 | 24 | 22 | |
| Glencoe 1 | 6 | 6 | 7 | 20 | 25 | 8 | 18 | 23 | 10 | 3 | 25 | 25 | |
| Glencoe 2 | 6 | 6 | 0 | 14 | 18 | 6 | 15 | 20 | 9 | 6 | 18 | 20 | |
| Glencoe 3 | 6 | 6 | 0 | 0 | 5 | 18 | 11 | 8 | 20 | 20 | 5 | 4 | |
| New Castle | 6 | 6 | 0 | 0 | 0 | 19 | 9 | 12 | 21 | 24 | 2 | 8 | |
| Oklahoma | 4 | 4 | 4 | 4 | 4 | 4 | 17 | 14 | 10 | 6 | 19 | 16 | |
| Oklahoma City | 2 | 2 | 6 | 6 | 6 | 6 | 6 | 9 | 18 | 16 | 10 | 11 | |
| Okmulgee | 7 | 7 | 1 | 1 | 1 | 1 | 5 | 5 | 22 | 22 | 12 | 10 | |
| Pawhuska | 0 | 0 | 6 | 6 | 6 | 6 | 4 | 2 | 7 | 10 | 20 | 20 | |
| Stillwater 1 | 6 | 6 | 0 | 0 | 0 | 0 | 4 | 6 | 1 | 6 | 24 | 24 | |
| Stillwater 2 | 1 | 1 | 7 | 7 | 7 | 7 | 5 | 1 | 6 | 1 | 7 | 8 | |
| Stigler | 7 | 7 | 1 | 1 | 1 | 1 | 3 | 7 | 2 | 7 | 1 | 8 | |
The numbers above the diagonal are for msp1α, and those below the diagonal are for msp4.
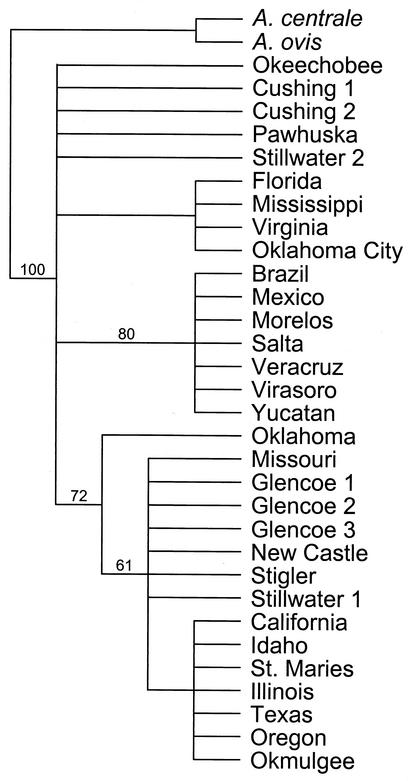
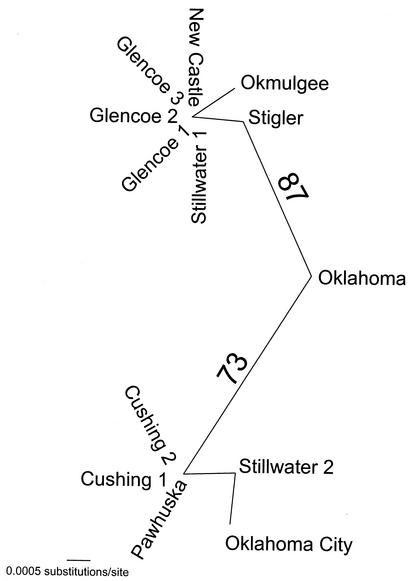
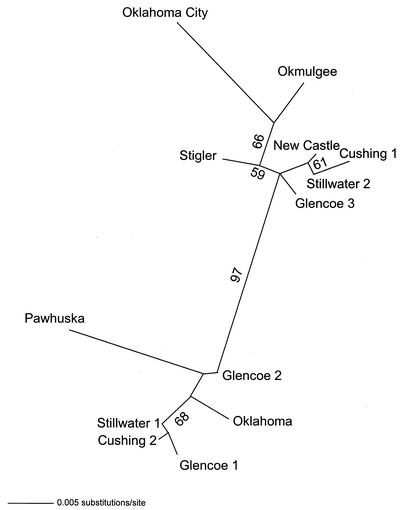
For the 31 msp4 sequences, only 39 characters were phylogenetically informative. MP analysis of these 39 characters resulted in 15 equally parsimonious trees of 51 steps (consistency index = 0.7843; retention index = 0.9209). Strong support was detected for a Latin American clade and a clade of isolates from the west-central United States (Fig. 3). Isolates from Oklahoma did not cluster together (Fig. 3). When only the msp4 sequences from Oklahoma isolates were considered, MODELTEST chose the modification of the GTR + I + Γ (where GTR is general time reversible, I is the proportion of invariable sites, and Γ is the shape parameter of the gamma distribution) model of DNA sequence evolution as most appropriate. The model parameters used in the ML analysis that resulted in a single tree were as follows: base frequencies = 0.25, 0.25, and 0.25; NST (number of substitution types) = 6; Revmat (rate matrix) = 1.0000, 16.9376, 1.0000, 1.0000, and 1.0000; rates = gamma; alpha parameter of the gamma distribution (α) = 0.9197; and proportion of invariant sites (Pinvar) = 0.939 (Fig. 4). Strong bootstrap analysis support (73%) was detected for east-central (isolates Oklahoma, Stigler, Okmulgee, New Castle, and Glencoe 1 to 3) and north-central (isolates Oklahoma City, Cushing 1 and 2, and Pawhuska) clades within Oklahoma (Fig. 4). Both clades contained isolates from Stillwater (Fig. 4).
FIG. 3.
Topology of the strict consensus of 15 equally parsimonious trees of 51 steps (consistency index = 0.7843; retention index = 0.9209) based on unweighted MP analysis of msp4 sequence data. The numbers above the branches indicate the percent bootstrap analysis support for those clades supported in more than 50% of 200 bootstrap iterations with 25 random additions of input taxa and TBR branch swapping.
FIG. 4.
Unrooted phylogram obtained by ML analysis depicting the phylogenetic relationships among Oklahoma A. marginale isolates based on msp4 sequence data. The numbers above the branches indicate the percent bootstrap analysis support for those clades supported in more than 50% of 200 bootstrap iterations.
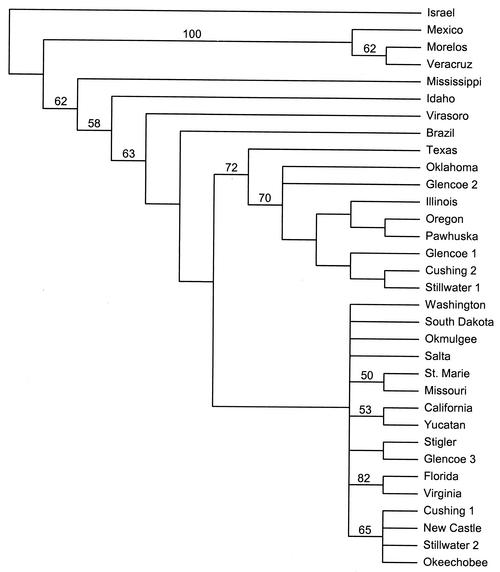
In contrast to the results obtained for msp4, MP analysis of the msp1α sequences demonstrated greater variability, which resulted in less phylogenetic and phylogeographic resolution than that afforded by use of msp4 sequences. The msp1α sequences of New World A. marginale isolates and one isolate from Israel provided 72 phylogenetically informative characters. MP analysis of these data resulted in 603 equally parsimonious trees of 163 steps (consistency index = 0.4908; retention index = 0.7552) and little support for phylogeographic clustering with the exception of a clade containing the isolates from Morelos and Veracruz, Mexico (Fig. 5). MODELTEST chose a modification of the GTR + I + Γ model of DNA substitution as the one that best fit the evolution of msp1α sequences of Oklahoma A. marginale isolates. The model parameters used in the ML analysis that resulted in a single tree were as follows: base frequencies = 0.2311, 0.2011, and 0.2986; NST = 6; Revmat = 1.0000, 7.8706, 1.0000, 1.0000, and 3.0680; rates = gamma; α = 0.7835; and Pinvar = 0.7001 (Fig. 6). In agreement with the results of MP analysis, ML analysis found little support for any phylogeographic structuring of the msp1α sequences of isolates from Oklahoma (Fig. 6). Similar results were obtained by MP and ML analyses of the MSP1a protein sequence data set (data not shown).
FIG. 5.
Topology of the strict consensus of 630 equally parsimonious trees of 163 steps (consistency index = 0.4908; retention index = 0.7552) based on unweighted MP analysis of msp1α sequence data. The numbers above the branches indicate the percent bootstrap analysis support for those clades supported in more than 50% of 200 bootstrap iterations with 25 random additions of input taxa and TBR branch swapping.
FIG. 6.
Unrooted phylogram obtained by ML analysis depicting the phylogenetic relationships among Oklahoma A. marginale isolates based on msp1α sequence data. The numbers above the branches indicate the percent bootstrap analysis support for those clades supported in more than 50% of 200 bootstrap iterations.
The ratio of the mean dN/dS can be used as an indicator of the level of selection acting on protein-coding genes. For msp4, dSs (dS = 0.0176 ± 0.0083) occurred at a frequency greater than dNs (dN = 0.0015 ± 0.0010), producing a dN/dS of 0.0852. For msp1α, dS was equal to 0.0286 ± 0.0157, whereas dN was equal to 0.0468 ± 0.0119, resulting in a dN/dS of 1.636. Thus, the selective pressures acting on msp1α are moderately positive and differ from those acting on msp4.
DISCUSSION
A. marginale geographic isolates have been defined by the sequence and number of repeats of MSP1a (1, 6, 9). In a previous study of isolates of A. marginale from the United States, we showed that MSP1a and MSP4 DNA and protein sequences provide phylogeographic information when the sequences of both are analyzed together (6). However, on a broader geographic scale, analysis of New World isolates demonstrated that msp4 sequences, but not MSP1a DNA or protein sequences, provide phylogeographic information (9). These results, together with the finding that multiple msp1α genotypes circulate in some geographic regions (e.g., an area of endemicity in Oregon [23]), questioned the use of MSP1α sequences for identification of geographic isolates of A. marginale and suggested that msp1α sequences may be rapidly evolving (9). In this study we tested these hypotheses by analyzing the msp1α and msp4 genotypes of A. marginale isolates obtained during 2001from infected cattle in east-central Oklahoma, where A. marginale is endemic, and comparing them with those of New World isolates.
As in previous studies (6, 9), msp4 sequences provided phylogeographic patterns for A. marginale isolates on a broad geographic scale. Additionally, DNA sequence characters further subdivided the U.S. isolates into west-central and southeastern clades. Both clades contained isolates from Oklahoma, suggesting extensive movement of cattle. Cattle movement during the 19th and early 20th centuries was responsible for the dissemination of tick-borne diseases in the United States (29, 30). On a smaller geographic scale with only the Oklahoma isolates, the msp4 sequences provided some phylogeographic information. However, the east-central and north-central clades included isolates from Stillwater, again suggesting the movement of cattle across this area. In contrast, analysis of the MSP1a DNA and protein sequences demonstrated extensive genotypic variation among Oklahoma isolates of A. marginale and did not provide phylogeographic information.
These results suggest that even if msp1α sequences are rapidly evolving, msp1α genotypes probably reflect the history of cattle movement rather than the geographic distribution of A. marginale isolates. Recent results suggest that different A. marginale genotypes are maintained within a herd in an area of endemicity by independent transmission events and that infection with more than one genotype per host is prevented by a mechanism of infection exclusion (10, 23). Therefore, if cattle movement introduces a new A. marginale genotype, it could be established by mechanical and/or biological transmission to susceptible cattle. In regions with few introductions of A. marginale isolates such as Australia, genotypic variation was found to be minimal (3, 19). In regions like Oklahoma, where the movement of cattle is extensive, a highly heterogeneous population of A. marginale isolates would be expected.
MSP1a is an immunodominant surface ligand involved in the interaction of A. marginale with host cell receptors (5, 7, 8). Furthermore, the tandem repeats that constitute the variable region of the protein mediate the interactions of the rickettsia with host cells and codify the capacity of the isolate to be transmitted by Dermacentor ticks (5, 8, 11). The support for positive selection pressure on msp1α is consistent with the biological function of MSP1a and suggests that this protein could be useful as a vaccine candidate. The sequence of MSP4 is highly conserved among A. marginale isolates and between A. marginale isolates and other Anaplasma spp. (9), but its biological function is unknown. Therefore, the lack of support for positive selection pressure on msp4 is difficult to interpret at present. Recently, Allsopp et al. (2) obtained similar results for the major antigenic protein (map1) of Ehrlichia ruminantium, which shows sequence identity with msp4, and suggested that map1 may not be a good candidate antigen for vaccination against E. ruminantium.
In summary, as we have demonstrated in previous studies (6, 9), msp4 sequences provided sufficient variation to detect the phylogeographic patterns of A. marginale isolates on different geographic scales. The results support the use of msp4 as a marker for elucidation of the phylogeographic patterns and the phylogenetic relationships among A. marginale isolates. In contrast to the results obtained with msp4, the DNA and deduced amino acid sequences of MSP1a failed to resolve either the phylogenetic or phylogeographic patterns of A. marginale isolates. The results indicate that MSP1a is not a marker for the characterization of geographic isolates of A. marginale and suggest that cattle movement and the maintenance of different genotypes by independent transmission events could explain the genetic heterogeneity observed among isolates of A. marginale in Oklahoma and, possibly, in other regions of endemicity.
Acknowledgments
This research was supported by project 1669 of the Oklahoma Agricultural Experiment Station, the Endowed Chair for Food Animal Research (K. M. Kocan, College of Veterinary Medicine, Oklahoma State University), the NIH Centers for Biomedical Research Excellence through a subcontract to J. de la Fuente from the Oklahoma Medical Research Foundation, and the Oklahoma Center for the Advancement of Science and Technology (applied research grant AR00(1)-001).
Yu-Wei Chiang and Stephen Whu (Fort Dodge Animal Health, American Home Products, Fort Dodge, Iowa) and Katherine Curtis (University of Missouri) are acknowledged for providing the A. marginale Pawhuska and Missouri isolates, respectively. The contributions of Robert Mihalovich (Large Animal Resident, Oklahoma State University), field veterinarians, and cattle farmers in the collection of blood samples from infected cattle in Oklahoma are acknowledged. Dollie Clawson (Department of Veterinary Pathobiology, Oklahoma State University) is acknowledged for technical assistance. Sue Ann Hudiburg and Janet J. Rogers (Core Sequencing Facility, Department of Biochemistry and Molecular Biology, Noble Research Center, Oklahoma State University) are acknowledged for oligonucleotide synthesis and DNA sequencing, respectively.
REFERENCES
- 1.Allred, D. R., T. C. McGuire, G. H. Palmer, S. R. Leib, T. M. Harkins, T. F. McElwain, and A. F. Barbet. 1990. Molecular basis for surface antigen size polymorphisms and conservation of a neutralization-sensitive epitope in Anaplasma marginale. Proc. Natl. Acad. Sci. USA 87:3220-3224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Allsopp, M. T., C. M. Dorfling, J. C. Maillard, A. Bensaid, D. T. Haydon, H. van Heerden, and B. A. Allsopp. 2001. Ehrlichia ruminantium major antigenic protein gene (map1) variants are not geographically constrained and show no evidence of having evolved under positive selection pressure. J. Clin. Microbiol. 39:4200-4203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bock, R. E., and A. J. de Vos. 2001. Immunity following use of Australian tick fever vaccine: a review of the evidence. Aust. Vet. J. 79:832-839. [DOI] [PubMed] [Google Scholar]
- 4.Bowie, M. V., J. de la Fuente, K. M. Kocan, E. F. Blouin, and A. F. Barbet. 2002. Conservation of major surface protein 1 genes of the ehrlichial pathogen Anaplasma marginale during cyclic transmission between ticks and cattle. Gene 282:95-102. [DOI] [PubMed] [Google Scholar]
- 5.de la Fuente, J., J. C. Garcia-Garcia, E. F. Blouin, S. D. Rodríguez, M. A. García, and K. M. Kocan. 2001. Evolution and function of tandem repeats in the major surface protein 1a of the ehrlichial pathogen Anaplasma marginale. Anim. Health Res. Rev. 2:163-173. [PubMed] [Google Scholar]
- 6.de la Fuente, J., R. A. Van Den Bussche, and K. M. Kocan. 2001. Molecular phylogeny and biogeography of North American isolates of Anaplasma marginale (Rickettsiaceae: Ehrlichieae). Vet. Parasitol. 97:65-76. [DOI] [PubMed] [Google Scholar]
- 7.de la Fuente, J., J. C. Garcia-Garcia, E. F. Blouin, and K. M. Kocan. 2001. Differential adhesion of major surface proteins 1a and 1b of the ehrlichial cattle pathogen Anaplasma marginale to bovine erythrocytes and tick cells. Int. J. Parasitol. 31:145-153. [DOI] [PubMed] [Google Scholar]
- 8.de la Fuente, J., J. C. Garcia-Garcia, E. F. Blouin, and K. M. Kocan. 2001. Major surface protein 1a effects tick infection and transmission of the ehrlichial pathogen Anaplasma marginale. Int. J. Parasitol. 31:1705-1714. [DOI] [PubMed] [Google Scholar]
- 9.de la Fuente. J., R. A. Van Den Bussche, J. C. Garcia-Garcia, S. D. Rodriquez, M. A. Garcia, A. A. Guglielmone, A. J. Mangold, L. M. Friche Passos, M. F. Barbosa Ribeiro, E. F. Blouin, and K. M. Kocan. 2002. Phylogeography of New World isolates of Anaplasma marginale (Rickettsiaceae: Anaplasmataceae) based on major surface protein sequences. Vet. Microbiol. 88:275-285. [DOI] [PubMed] [Google Scholar]
- 10.de la Fuente, J., J. C. Garcia-Garcia, E. F. Blouin, J. T. Saliki, and K. M. Kocan. 2002. Infection of tick cells and bovine erythrocytes with one genotype of the intracellular ehrlichia Anaplasma marginale excludes infection with other genotypes. Clin. Diagn. Lab. Immunol. 9:658-668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.de la Fuente, J., J. C. Garcia-Garcia, E. F. Blouin, and K. M. Kocan. 2003. Characterization of the functional domain of major surface protein 1a involved in adhesion of the rickettsia Anaplasma marginale to host cells. Vet. Microbiol. 91:265-283. [DOI] [PubMed] [Google Scholar]
- 12.Dumler, J. S., A. F. Barbet, C. P. J. Bekker, G. A. Dasch, G. H. Palmer, S. C. Ray, Y. Rikihisa, and F. R. Rurangirwa. 2001. Reorganization of the genera in the families Rickettsiaceae and Anaplasmataceae in the order Rickettsiales: unification of some species of Ehrlichia with Anaplasma, Cowdria with Ehrlichia and Ehrlichia with Neorickettsia, descriptions of six new species combinations and designation of Ehrlichia equi and “HGE agent” as subjective synonyms of Ehrlichia phagocytophila. Int. J. Syst. E vol. Microbiol. 51:2145-2165. [DOI] [PubMed] [Google Scholar]
- 13.Ewing, S. A. 1981. Transmission of Anaplasma marginale by arthropods, p. 395-423. In Proceedings of the 7th National Anaplasmosis Conference. Mississippi State University, Mississippi State.
- 14.Felsenstein, J. 1985. Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:783-791. [DOI] [PubMed] [Google Scholar]
- 15.Jukes, T. H., and C. R. Cantor. 1969. Evolution of protein molecules, p. 21-132. In H. N. Munro (ed.), Mammalian protein metabolism. Academic Press, Inc., New York, N.Y.
- 16.Kano, F. S., O. Vidotto, R. C. Pacheco, and M. C. Vidotto. 2002. Antigenic characterization of Anaplasma marginale isolates from different regions of Brazil. Vet. Microbiol. 87:131-138. [DOI] [PubMed] [Google Scholar]
- 17.Kocan, K. M., T. Halbur, E. F. Blouin, V. Onet, J. de la Fuente, J. C. Garcia-Garcia, and J. T. Saliki. 2001. Immunization of cattle with Anaplasma marginale derived from tick cell culture. Vet. Parasitol. 102:151-161. [DOI] [PubMed] [Google Scholar]
- 18.Kumar, S., K. Tamura, and M. Nei. 1994. MEGA: Molecular Evolutionary Genetics Analysis software for microcomputers. Comput. Appl. Biosci. 10:189-191. [DOI] [PubMed] [Google Scholar]
- 19.Lew, A. E., R. E. Bock, C. M. Minchin, and S. Masaka. 2002. A msp1α polymerase chain reaction assay for specific detection and differentiation of Anaplasma marginale isolates. Vet. Microbiol. 86:325-335. [DOI] [PubMed] [Google Scholar]
- 20.McGarey, D. J., and D. R. Allred. 1994. Characterization of hemagglutinating components on the Anaplasma marginale initial body surface and identification of possible adhesins. Infect. Immun. 62:4587-4593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McGarey, D. J., A. F. Barbet, G. H. Palmer, T. C. McGuire, and D. R. Allred. 1994. Putative adhesins of Anaplasma marginale: major surface polypeptides 1a and 1b. Infect. Immun. 62:4594-4601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nei, M., and T. Gojobori. 1986. Simple methods for estimating the numbers of synonymous and nonsynonymous nucleotide substitutions. Mol. Biol. Evol. 3:418-426. [DOI] [PubMed] [Google Scholar]
- 23.Palmer, G. H., F. R. Rurangirwa, and T. F. McElwain. 2001. Strain composition of the ehrlichia Anaplasma marginale within persistently infected cattle, a mammalian reservoir for tick transmission. J. Clin. Microbiol. 39:631-635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Posada, D., and K. A. Crandall. 1998. MODELTEST: testing the model of DNA substitution. Bioinformatics 14:817-818. [DOI] [PubMed] [Google Scholar]
- 25.Ristic, M., and A. M. Watrach. 1963. Anaplasmosis. VI. Studies and a hypothesis concerning the cycle of development of the causative agent. Am. J. Vet. Res. 24:267-276. [PubMed] [Google Scholar]
- 26.Saitou, N., and M. Nei. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406-425. [DOI] [PubMed] [Google Scholar]
- 27.Swofford, D. L. 2000. PAUP* phylogenetic analysis using parsimony (*and other methods) version 4.02b. Sinauer Associates, Inc., Publishers, Sunderland, Mass.
- 28.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, positions-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.U.S. Department of Agriculture. 1893. Eight and ninth annual reports of the Bureau of Animal Industry for the years 1891 and 1892, p. 25-28. Government Printing Office, Washington, D.C.
- 30.U.S. Department of Agriculture. 1912. Twenty-seventh annual report of the Bureau of Animal Industry for the year 1910, p. 535-543. Government Printing Office, Washington, D.C.