Abstract
Nitrous oxide (N2O) is an important greenhouse gas in the troposphere controlling ozone concentration in the stratosphere through nitric oxide production. In order to quantify bacteria capable of N2O reduction, we developed a SYBR green quantitative real-time PCR assay targeting the nosZ gene encoding the catalytic subunit of the nitrous oxide reductase. Two independent sets of nosZ primers flanking the nosZ fragment previously used in diversity studies were designed and tested (K. Kloos, A. Mergel, C. Rösch, and H. Bothe, Aust. J. Plant Physiol. 28:991-998, 2001). The utility of these real-time PCR assays was demonstrated by quantifying the nosZ gene present in six different soils. Detection limits were between 101 and 102 target molecules per reaction for all assays. Sequence analysis of 128 cloned quantitative PCR products confirmed the specificity of the designed primers. The abundance of nosZ genes ranged from 105 to 107 target copies g−1 of dry soil, whereas genes for 16S rRNA were found at 108 to 109 target copies g−1 of dry soil. The abundance of narG and nirK genes was within the upper and lower limits of the 16S rRNA and nosZ gene copy numbers. The two sets of nosZ primers gave similar gene copy numbers for all tested soils. The maximum abundance of nosZ and nirK relative to 16S rRNA was 5 to 6%, confirming the low proportion of denitrifiers to total bacteria in soils.
Nitrous oxide (N2O), with a global warming potential approximately 300 times higher than that of carbon dioxide, is an important greenhouse gas, contributing up to 6% of global warming. N2O also participates in depletion of the stratospheric ozone layer through stratospheric nitric oxide (NO) production. At present, the N2O concentration in the atmosphere is increasing at a rate of about 0.3% per year. The soil is the dominant source of atmospheric nitrous oxide, contributing about 57% (9 Tg year−1) of the total annual global emission (12).
N2O emissions are highly variable in soils and are primarily produced by biological nitrification and denitrification, although the latter is considered to be the main source (31). N2O is an intermediate product in the denitrification pathway, which consists of the sequential reduction of NO3− to N2 via the metalloenzymes nitrate reductase, nitrite reductase, nitric oxide reductase, and nitrous oxide reductase (32). Therefore, N2O emission by denitrification is the net result of the balance between production and reduction of N2O by denitrifying bacteria. The N2O reductase (EC 1.7.99.6) is a homodimeric multicopper enzyme, which has been purified from numerous gram-negative denitrifiers but not yet from a gram-positive bacterium (4, 5, 11, 17, 27). Production of N2O by denitrifying isolates as an end product of denitrification has been reported by several authors (2, 3, 7). Sequencing of the complete genome of Agrobacterium tumefaciens C58 revealed the presence of a denitrification cluster with genes encoding the periplasmic nitrate reductase, the copper nitrite reductase, and the nitric oxide reductase, but the genes encoding the N2O reductase were not found within the entire genome (30). These results suggest that N2O production by denitrifiers may be due not only to the regulation of nitrous oxide reductase activity but also to an absence of genes encoding this enzyme in some denitrifying populations.
Since an unknown percentage of denitrifying bacteria lack the genes encoding N2O reductase and will emit N2O as an end product in denitrifying conditions, irrespective of soil physicochemical characteristics, it is important to know the abundance of bacteria able to reduce N2O in order to better understand the key drivers of N2O emissions from soil. Recent papers have underlined the potential role of the composition of the denitrifying community among the factors leading to N2O emission (1, 9).
In the present study, we report the development of a quantitative PCR assay to quantify the nosZ gene encoding the catalytic subunit of N2O reductase. The specificity and sensitivity of the assay were investigated with environmental samples, and the results were compared with those of the real-time quantification of the 16S rRNA, narG (encoding the membrane-bound nitrate reductase), and nirK (encoding the copper nitrite reductase) genes.
MATERIALS AND METHODS
Environmental samples and bacterial strains.
Soil samples were taken from Ljubljana marsh (Slovenia), two agricultural soils located in Champ Noël and Nancy (France), and three soils from the northeastern Himalayas (Nepal). All six soil samples were taken at a depth of 5 to 10 cm, and their characteristics are described in Table 1. Total soil nitrogen was measured as Kjeldahl nitrogen (International Organization for Standardization [ISO] standard 10694). The contents of organic carbon were determined through dry combustion (ISO standard 13878), and the pH was defined in a solution of 0.01 M calcium chloride (ISO standard 10390). The bacterial isolates used for the specificity studies are listed in Table 2.
TABLE 1.
Properties of the soils analyzed
| Soil | % of component
|
Concn of element (mg/kg)
|
pH | |||
|---|---|---|---|---|---|---|
| Clay | Sand | Silt | Nitrogen | Organic carbon | ||
| Ljubljana marsh | 6 | 16.3 | 77.7 | 1.3 | 150 | 6 |
| Himalaya | ||||||
| 6,000 m | 0.5 | 69.5 | 30 | 0.50 | 3.73 | 5.41 |
| 5,600 m | 0 | 80 | 20 | 0.85 | 8.72 | 5.89 |
| 5,400 m | 0 | 68 | 32 | 0.84 | 6.07 | 5.89 |
| Champ Noël | 14.1 | 19.3 | 66.6 | 1.04 | 9.4 | 5.89 |
| Nancy | 33.3 | 15.4 | 51.3 | 1.7 | 15.3 | 5.8 |
TABLE 2.
Bacterial strains used in this study to test specificity of the nosZ1 and nosZ2 primers
| Strain | Group | Presence of nosZ | N2O reduction | Quantitative PCR result with primer type:
|
|
|---|---|---|---|---|---|
| nosZ1 | nosZ2 | ||||
| Bacillus senegalensis M1518 | Firmicutes | NDb | + | − | − |
| Bacillus cereus M944 | Firmicutes | ND | + | − | − |
| Bacillus licheniformis SN530 | Firmicutes | ND | − | − | − |
| Bradyrhizobium japonicum 562 | α-Proteoa | + | ND | + | − |
| Bradyrhizobium japonicum USDA110 | α-Proteo | + | ND | + | + |
| Sinorhizobium morelense SN611 | α-Proteo | + | ND | + | + |
| Hyphomicrobium denitrificans DSM 1869 | α-Proteo | + | ND | + | − |
| Sinorhizobium meliloti 1021 | α-Proteo | + | ND | + | + |
| Rhizobium meliloti 50 | α-Proteo | + | ND | + | + |
| Alcaligenes faecalis ATCC 8750 | β-Proteo | + | ND | + | + |
| Achromobacter cycloclastes ATCC 21921 | β-Proteo | + | ND | + | + |
| Achromobacter xylosoxidans M129 | β-Proteo | + | + | + | + |
| Pseudomonas denitrificans CCUG 2519 | γ-Proteo | + | ND | + | − |
| Pseudomonas fluorescens C7R12 | γ-Proteo | + | + | + | + |
| Escherichia coli JM109 | γ-Proteo | − | − | − | − |
Proteo, proteobacteria.
ND, not determined.
DNA extraction.
DNA was extracted from three 250-mg aliquots of soil samples (16). Briefly, samples were homogenized in 1 ml of extraction buffer (Tris-HCl, 100 mM, pH 8; EDTA, 100 mM, pH 8; NaCl, 100 mM; 1% polyvinylpolypyrrolidone; 2% sodium dodecyl sulfate) for 30 s at 1,600 rpm in a Minibead-beater cell disrupter (Mikro-DismembratorS; B. Braun Biotech International). Soil and cell debris were eliminated by centrifugation (14,000 × g for 5 min at 4°C). The proteins were then removed with 3 M (pH 5.5) sodium acetate and the nucleic acids precipitated using cold isopropanol. The DNA was washed with 70% ethanol and purified through a Sepharose 4B spin column. Finally the DNA was quantified by spectrophotometry at 260 nm using a BioPhotometer (Eppendorf, Germany).
Primer design.
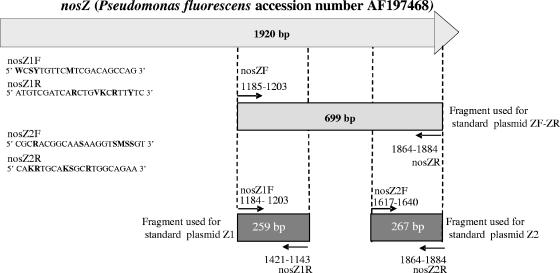
Two independent sets of nosZ primers were designed by aligning all publicly available complete nosZ sequences using the Fungene database (http://flyingcloud.cme.msu.edu/fungene/) and searching for conserved regions that could provide suitable primer target sites. Four degenerate primers, nosZ1F-nosZ1R and nosZ2F-nosZ2R, were selected to amplify a 259- and a 267-bp fragment, respectively, within the region amplified using the diversity primers nosZF-nosZR designed by Kloos et al. (13) (Fig. 1). The specificity of the primers was tested in silico using the Fungene software (http://flyingcloud.cme.msu.edu/fungene/) and 42 complete nosZ sequences from known strains. In addition, 15 strains were also used to evaluate primer specificity.
FIG. 1.
Sequences and binding positions of the nosZ1 and nosZ2 primers based on the nosZ sequence from Pseudomonas fluorescens (accession number AF197478). Degenerated sites are in boldface (W = AT, S = CG, Y = CT, M = AC, R = AG, K = GT, and V = ACG).
Standard curves.
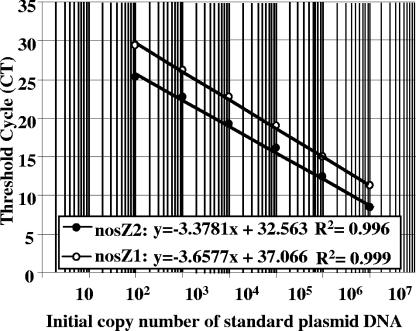
The three plasmids containing nosZ gene fragments amplified from Pseudomonas fluorescens C7R12 with the nosZ1, nosZ2, and nosZF-nosZR primers were used as standards (Fig. 1). The nosZ PCR products were cloned using the pGEM-T Easy cloning kit according to the manufacturer's instructions (Promega, France). The recombinant Escherichia coli JM109 strains carrying the nosZ recombinant plasmids were inoculated into LB broth with ampicillin (100 μg liter−1) and incubated at 37°C overnight. Plasmid DNA was then extracted using the MiniPrep kit from QIAGEN (France) according to the manufacturer's instructions, and the plasmid concentrations were determined by spectrophotometry using a BioPhotometer (Eppendorf, Germany). Plasmids containing the nosZ1 and nosZ2 fragments were linearized using the EcoRI restriction enzyme, while the plasmid containing the 700-bp nosZF-nosZR fragment was digested with SalI. The presence of nosZ inserts was verified by gel electrophoresis. Standards were prepared from linearized plasmid serial dilutions containing between 107 and 102 nosZ copies calculated directly from the concentration of extracted plasmid. The threshold cycle, the cycle at which the fluorescence in the sample increases above a defined threshold, is inversely proportional to the starting amount of nucleic acid (Fig. 2). Standard curves were generated by plotting the threshold cycle for each standard, calculated according to the ABI Prism 7900 software, against the log10 of the gene copy number. The amplification efficiency (E) was estimated by using the slope of the standard curve according to the following formula: E = (10−1/slope) − 1.
FIG. 2.
Standard curves of the nosZ1 and nosZ2 assays obtained by plotting the concentration of control DNA versus the cycle number required to elevate the fluorescence signal above the threshold.
Quantitative PCR assays.
The real-time PCR assay was carried out in a volume of 25 μl, and the assay mixture contained SYBR green PCR Master Mix (QuantiTect SYBR green PCR kit; QIAGEN, France), 1 μM of each nosZ primer, 100 ng of T4 gp 32 (QBiogene, France), and 1.25 μl of template DNA (at 2, 5, or 10 ng μl−1). Thermal cycling conditions for the nosZ1 primers were as follows: an initial cycle of 95°C for 15 min; 6 cycles of 95°C for 15 s, 67°C for 30 s with a touchdown of −1°C by cycle, 72°C for 30 s, and 80°C for 15 s (acquisition data step); 40 cycles of 95°C for 15 s and 62°C for 15 s, 72°C for 30 s, and 80°C for 15 s; and 1 cycle at 95°C for 15 s and 60°C for 15 s, to 95°C for 15 s. The thermal cycling conditions for the nosZ2 primers were similar except for the annealing temperature starting at 65°C for the first 6 cycles and 60°C for the 40 cycles. Thermal cycling, fluorescent data collection, and data analysis were carried out with the ABI Prism 7900 sequence detection system according to the manufacturer's instructions. Quantitative PCR assays of the 16S rRNA, narG, and nirK genes were performed as described previously (8, 15). Two independent quantitative PCR assays were performed for each gene and for the three replicate DNA extractions from each soil sample. Two or three no-template controls (NTCs) were run for each quantitative PCR assay. All 16S rRNA, nirK, and nosZ assays were also run with DNA from Bradyrhizobium japonicum USDA110 containing one copy of each of these three genes.
DNA extracts from all soils which exhibit nosZ copy numbers ranging between 102 and 103 copies per ng were spiked with the plasmid DNA at a concentration of 104 to 105 copies per ng, and gene copy numbers were compared to those in samples without soil DNA to test for the potential presence of PCR inhibitors. In addition, serial dilutions of the DNA extracted from soil samples were quantified and compared.
Construction of nosZ quantitative PCR product clone libraries and phylogenetic analysis.
The specificity of the assays was verified by constructing eight clone libraries using the nosZ1 and nosZ2 real-time PCR products from DNA extracted from the Nancy, Champ Noël, Himalaya (6,000-m), and Ljubljana soils. The three replicates of quantitative PCR products from each soil were pooled and cloned into the plasmid vector pGEM-T Easy according to the manufacturer's instructions (Promega, France). Cells were spread onto LB agar plates containing ampicillin and grown overnight at 37°C. The colonies were then randomly picked, transferred to new plates, and incubated overnight at 37°C. Small aliquots of cells from each colony were transferred to a PCR mixture containing the vector primers T7 and SP6. Clones containing the insert DNA were identified by agarose gel electrophoresis. Nucleotide sequences of the inserted quantitative PCR products were determined using the GenomeLab DTCS Quick Start kit (Beckman Coulter) with primer T7 in a Ceq 8000 sequencer, according to the manufacturer's instructions (Beckman Coulter).
One hundred twenty-eight nosZ sequences from the eight clone libraries were deposited in GenBank (accession numbers given below). Alignments of nosZ fragments of 259 and 267 nucleotides from quantitative PCR products and from reference strains were made by using the CLUSTAL X software version 1.0.1. The neighbor-joining method was used to calculate the distances and to construct phylogenic trees.
Data analysis.
For each soil replicate, quantitative PCR was performed twice for each gene. Replicate results of quantitative PCR measurements were averaged (n = 6), and the standard error was calculated. Comparison of the gene copy numbers within the same soil and between soils was performed using the Fisher test in StatView version 5.0 (SAS Institute Inc.).
Nucleotide sequence accession numbers.
The 128 nosZ sequences from the eight clone libraries were deposited in GenBank under accession numbers AJ786667 to AJ786710.
RESULTS
Primer specificity.
In silico evaluation of the nosZ1 and nosZ2 primers showed that 83, 71, 68, and 98% of the 42 complete nosZ sequences analyzed exhibited a maximum of one mismatch with the nosZ1F, nosZ1R, nosZ2F, and nosZ2R primers, respectively. The specificity of the nosZ1 and nosZ2 sets of primers was further verified experimentally by using phylogenetically diverse denitrifier strains (Table 3). Amplification with the nosZ1 primers resulted in the expected 259-bp fragment for all gram-negative strains except those of E. coli. In contrast, no products were amplified from the negative-control E. coli or from Bradyrhizobium japonicum 562, Hyphomicrobium denitrificans, and Pseudomonas denitrificans using the nosZ2 primers. In addition, for both sets of primers, no products were amplified from the Bacillus strains, which are capable of reducing N2O.
TABLE 3.
narG, nirK, and nosZ (obtained with nosZ1 and nosZ2 assays) calculated percentages of 16S rRNA copy numbers based on the gene copy number per ng of DNA
| Soil | Denitrification gene (% of 16S rRNA)
|
|||
|---|---|---|---|---|
| narG | nirK | nosZ1 | nosZ2 | |
| Nancy | 40 | 6 | 3 | 5 |
| Himalaya | ||||
| 6,000 m | 0.9 | 0.7 | 0.1 | 0.1 |
| 5,600 m | 1 | 0.8 | 0.1 | 0.1 |
| 5,400 m | 0.6 | 1.3 | 0.1 | 0.1 |
| Ljubljana marsh | 20 | 1.2 | 0.5 | 0.5 |
| Champ Nöel | 6 | 6 | 0.2 | 0.4 |
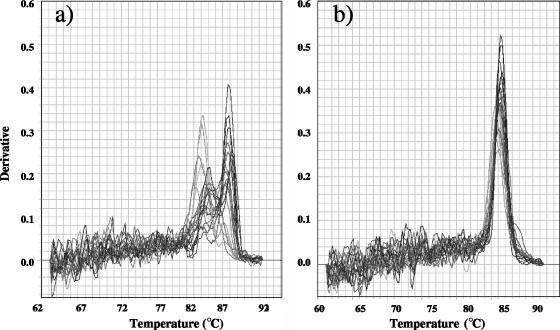
Melting-curve analysis with target DNA from strains (data not shown) or from environmental samples showed contrasting results between the nosZ1 and nosZ2 assays (Fig. 3). The first derivative of the melting curves of the nosZ2 assay shows one distinct peak (Fig. 3b), while several peaks can be seen for the nosZ1 assay (Fig. 3a). However, agarose gel electrophoresis showed the presence of only one band of the expected size for both nosZ1 and nosZ2 (data not shown). The specificity of the assays was further confirmed by cloning the quantitative PCR products from the 6,000-m Himalaya soil and for all the other soils. Sequencing of 59 and 69 cloned quantitative PCR products from the nosZ1 and nosZ2 assays, respectively, revealed the presence of one nonspecific PCR product for the nosZ2 assay and none for the nosZ1 assay. All other quantitative PCR products exhibited identities ranging between 61 and 85% and 68 and 88%, respectively, for nosZ1 and nosZ2 with the closest nosZ gene from a cultivated bacterium. In this respect, the presence of multiple melting peaks for the nosZ1 assay was caused by differing melting points of different sequences within the mixed nosZ1 quantitative PCR product and not by nonspecific amplification.
FIG. 3.
Melting-curve profiles for the amplicons of environmental DNA obtained by nosZ1 (a) and nosZ2 (b) primer sets.
Production of standard curves.
Standard curves for quantitative PCR were obtained by preparing 10-fold dilutions of three plasmids containing nosZ1, nosZ2, or a 699-bp nosZ fragment amplified with the nosZR-nosZF primers developed by Kloos et al. (13). All standard curves had shown a linear range between 107 and 102 copies, with a slope of −3.677 and −3.378 for nosZ1 and nosZ2, respectively (Fig. 2). The calculated PCR efficiencies for the nosZ1 and nosZ2 assays were 86% and 97%, respectively. PCR efficiencies for the 16S rRNA, narG, and nirK assays were 104, 99, and 105%, respectively.
We then compared the 16S rRNA and nosZ gene copy numbers to check that the standard curves gave a good estimation of the latter, using genomic DNA from Bradyrhizobium japonicum USDA110 as the template. Quantification of the gene copy number in 12.5 ng of DNA from B. japonicum using the nosZ1, nosZ2, and 16S rRNA quantitative PCR assays gave the same copy number of approximately 106. The experimental number was in accordance with the theoretical number estimated using an average molecular mass of 660 for a base pair in double-stranded DNA, the number of 16S rRNA and nosZ gene copies in the B. japonicum genome (one each), the genome size of B. japonicum of 9,105,828 bp (genome accession number NC 004460), and the spectrophotometrically determined concentration of genomic DNA.
Detection limit.
In our SYBR green assay, it was possible to discriminate against the primer dimer fluorescence by acquiring data at a temperature of 80°C, which is above the melting point of these by-products. This temperature was determined and verified by melting-curve analysis (Fig. 3). The gene copy numbers in the NTCs were 0 and less than 10 copies for nosZ1 and nosZ2, respectively. Higher values of 102 copies per assay were observed for narG and 16S rRNA quantification. According to the NTC values, a detection limit of approximately 10 to 100 target molecules per assay was achieved for nosZ quantification, which corresponds roughly to 103 to 104 targets per gram of dry soil.
Quantification of the 16S rRNA, narG, nirK, and nosZ genes.
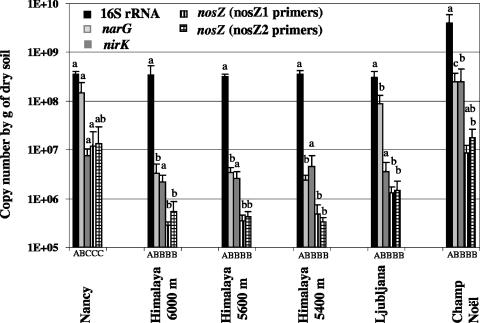
The applicability of the assay to quantify the nosZ gene in environmental samples was evaluated using DNA extracted from various soils as the template. No significant difference between the estimated nosZ gene copy numbers based on the nosZ1 or nosZ2 primer was recorded in any of the tested soils. The nosZ density in the Himalayan soil ranged from 2 × 105 to 5 × 105 copies g−1 of dry soil whatever the sampling site, whereas densities up to 107 copies g−1 of dry soil were observed in the French soils (Fig. 4). The reproducibility of two independent quantitative PCR assays with the same DNA extract was good, with average percentages of variation of 17% and 28% for nosZ1 and nosZ2, respectively (data not shown). The nirK gene density was between 106 and 107 copies g−1 of dry soil for all soils except that of Champ Noël, in which 2.5 × 108 copies g−1 of dry soil were quantified. This latter soil also exhibited significantly higher 16S rRNA and narG gene copy numbers (Fig. 4). The relative abundance of nosZ genes ranged from 0.1 to 0.5% of 16S rRNA genes, except in the soil from Nancy, where it was about 4% (Table 3). A slightly higher span of relative abundance (0.7 to 6% of 16S rRNA genes) was observed for the nirK genes.
FIG. 4.
16S rRNA, narG, nirK, and nosZ (obtained with nosZ1 and nosZ2 assays) copy numbers g−1 of dry soil in the different soils. Error bars indicate standard errors of the two independent PCRs of the three replicate DNA extractions. Significantly different values (P < 0.05) between different genes in the same soil are marked by capital letters (A to C) under the columns, and significantly different values (P < 0.05) between different soils for the same gene are marked by lowercase letters (a to c).
The presence of PCR inhibitors in the soil samples was tested by conducting a real-time PCR by spiking DNA extracts from all soils with control plasmid DNA. The signal fluorescence was the same for the control plasmid DNA both in the presence and in the absence of DNA extracted from the soil samples, thus indicating the absence of detectable inhibition in our assays. This was also confirmed by quantifying nosZ in serial dilutions of DNA extracted from the soil samples (data not shown).
Phylogenetic analysis of nosZ quantitative PCR products.
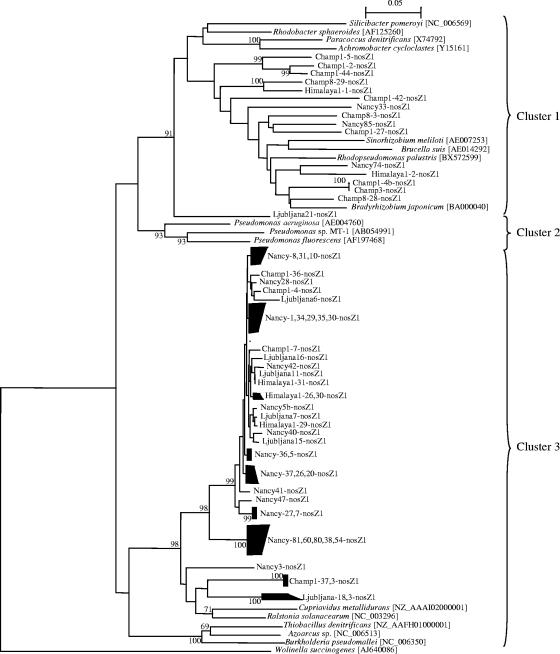
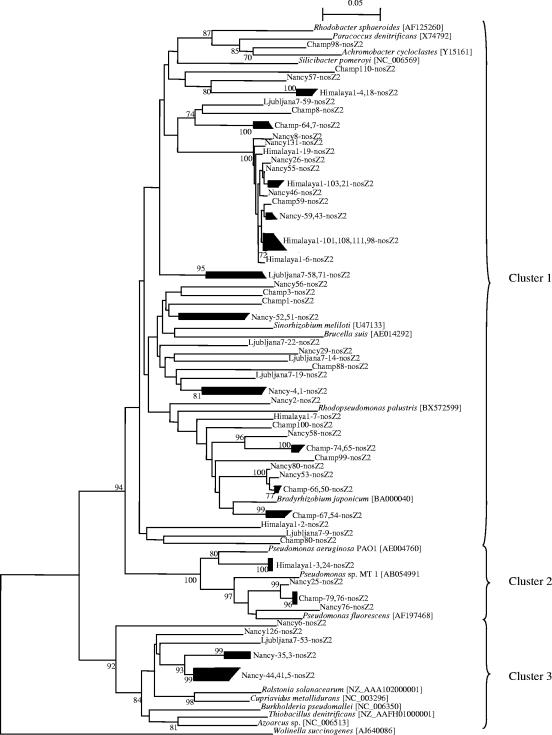
The applicability of the primers to environmental samples was verified by performing a phylogenetic analysis of the sequenced quantitative PCR products. The tree topology differed between the nosZ sequences from nosZ1 and nosZ2 sets of primers (Fig. 5 and 6). Analysis of the nosZ1 tree showed a large highly conserved group of sequences clustering next to the nosZ sequence from β-proteobacteria such as Ralstonia solanacearum and Cupriavidus metallidurans and a group of sequences close to nosZ genes from α-proteobacteria. Sequences obtained with the nosZ2 primers fell within three clusters, with most of the sequences clustering with nosZ from α-proteobacteria (Fig. 6). The second cluster encompassed sequences from all soils except Ljubljana and nosZ sequences from γ-proteobacteria such as Pseudomonas spp. The last cluster contained the sequences from Nancy and Ljubljana marsh and nosZ from β-proteobacteria.
FIG. 5.
Phylogenetic tree of nosZ quantitative PCR products obtained with the nosZ1 primers and of nosZ genes from reference strains. Fragments of 259 nucleotides from the quantitative PCR products and from reference strains were used to calculate the tree. The tree is based on distance matrix analysis and the neighbor-joining method. Bootstrap values greater than 700 from 1,000 replicate trees are reported at the nodes. The sequence of nosZ from Wolinella succinogenes served as an outgroup to root the tree.
FIG. 6.
Phylogenetic tree of nosZ quantitative PCR products obtained with the nosZ2 primers and of nosZ genes from reference strains. Fragments of 267 nucleotides from the quantitative PCR products and from reference strains were used to calculate the tree. The tree is based on distance matrix analysis and the neighbor-joining method. Bootstrap values greater than 700 from 1,000 replicate trees are reported at the nodes. The sequence of nosZ from Wolinella succinogenes served as an outgroup to root the tree.
DISCUSSION
Knowledge of the ecology of N2O-reducing bacteria is of crucial importance to better understand the factors governing N2O fluxes from the environment, especially from soils. Earlier studies demonstrated the role of the denitrifier community composition in N2O fluxes (1, 9). Although several authors reported the diversity of the N2O-reducing community (10, 18, 23, 24, 28), no studies were performed to estimate its actual density. The isolation and characterization of denitrifiers based on their N2O-reducing abilities (2) are hampered by cultivation-dependent bias and the extensive manual effort due to the fact that denitrifiers represent a low percentage of the total bacteria. In this study, an easy and rapid culture-independent real-time PCR assay was developed to quantify the nosZ genes encoding the catalytic subunit of the nitrous oxide reductase, and the results were compared with the numbers of 16S rRNA, narG, and nirK genes. SYBR green was used as the detection system, as it permits the application of a primer system to real-time PCR without developing further probes and has been successfully used to quantify other denitrification genes (20).
In order to relate with more accuracy the diversity and density data in future studies, quantitative PCR primers were designed in the regions flanking the nosZ fragment amplified for diversity analysis (13). In addition, two independent sets of primers, for nosZ1 and nosZ2, were used to verify the robustness of our assay. Since between 0 and fewer than 10 targets were detected in the no-template control, the detection limit of our nosZ assays was equivalent to 103 to 104 copies g−1 of dry soil, which was within the range of those previously reported (8, 14, 29). The standard curves were linear over 6 logs for both nosZ1 and nosZ2, with a very good correlation between the number of copies of target DNA and the cycle threshold value (Fig. 2). Quantification of nosZ in DNA from Bradyrhizobium japonicum USDA110 using either the nosZ1 or the nosZ2 set of primers gave no significant difference with the theoretical nosZ gene copy number of approximately 100,000 copies per ng of DNA, indicating that both the nosZ1 and nosZ2 assays can be used to accurately quantify nosZ. To further increase the robustness of the assays and facilitate the comparison of data obtained with the two sets of primers, three standard plasmids were constructed, one specific to each set of primers and one containing a 699-bp nosZ fragment that can be used for both sets of primers (Fig. 1).
Primer specificity was verified by amplifying a collection of denitrifiers and by sequencing the real-time PCR products from the soil samples. None of the nosZ primers tested in this study was able to amplify the Bacillus strains, indicating that our primers were probably specific only for the nitrous oxide reductase from gram-negative bacteria. In addition, nosZ sequences from a few strains, such as the ɛ-proteobacterium Wolinella succinogenes (25), exhibit too many mismatches with either the nosZ1 or nosZ2 primers to be successfully amplified. Analysis of the nosZ1 and nosZ2 phylogenetic trees showed different topologies, thereby confirming the differences in specificity between the two sets of nosZ primers. Most of the sequences amplified by nosZ1 clustered with nosZ from β-proteobacteria (Fig. 5), whereas the sequences obtained with the nosZ2 primers were more widely spread between clusters containing nosZ from α-proteobacteria and those containing nosZ from β-proteobacteria. Some sequences also clustered with nosZ from γ-proteobacteria (Fig. 6). A larger number of sequences was generated for the French soils from Nancy and Champ Noël to allow their comparison with sequences obtained by amplifying the same soils with the nosZ primers described by Kloos et al. (13; unpublished data). Comparison of the phylogenetic trees showed that sequences from the nosZ2 assay were more closely related to those obtained using the nosZ primer from Kloos et al. (13) than to those obtained using the nosZ1 primer (data not shown). Altogether, these data confirm that the diversity results obtained in PCR-based studies are strongly influenced by the initial choice of primer set. Analysis of the quantitative PCR products confirmed that at least the nosZ2 set of primers was capable of quantifying the nosZ gene from α-, β-, and γ-proteobacteria in soils. Based on the intuition that the primer set yielding the greater diversity would be more suitable, the nosZ2 primers seemed of greater interest than the nosZ1 primers. However, the superiority of either primer set cannot be confirmed since the real nosZ diversity in the studied soils (i.e., determined without any PCR or cultivation bias) is still unknown.
Although differences in specificity were observed between the nosZ1 and nosZ2 primers, similar nosZ1 and nosZ2 gene copy numbers were observed in each soil. Thus, application of the nosZ quantitative PCR assay to template DNA extracted from different soils showed that the numbers of nosZ genes ranged from 105 to 107 copies g−1 of dry soil (Fig. 4). The numbers of nirK copies were in the upper range of those found previously by Henry et al. (8) but 1 to 2 orders of magnitude lower than those observed by Qiu et al. using competitive PCR (22). Similar 16S rRNA gene copy numbers were also observed by Smith et al. (26). Up to now, only one copy of the nirK and nosZ genes has been observed in all the bacteria studied (19), suggesting that the nirK and nosZ gene copy numbers are directly correlated with cell numbers. Since denitrifiers generally contain either a copper nitrite reductase or a cytochrome cd1 nitrite reductase (32), the nosZ gene copy number is expected to be higher than the nirK gene copy number. However, quantification of nirK showed numbers on the same order of magnitude as nosZ or even 10 times higher in the soil from Champ Noël. This higher nirK gene copy number could be related to methodological bias, because nirK primers might be more universal than the nosZ primers. However, phylogenetic analysis of the sequenced quantitative PCR products showed that the nosZ fragments related to nosZ from α-, β-, and γ-proteobacteria were successfully amplified, which demonstrated that our primers are not restricted to a narrow group. Another hypothesis to explain these high nirK copy numbers is that a significant percentage of denitrifiers present in soil lack the nosZ gene, as has been demonstrated by sequencing the complete genome of Agrobacterium tumefaciens C58 (30). Therefore, it is tempting to suggest that the variations in nirK/nosZ ratio depending on the soil could be associated with differences in the proportion of denitrifiers lacking nosZ. In future studies, it will be interesting to test this hypothesis and its ecological consequences in terms of greenhouse gas emission by estimating the nirK- and/or nirS-to-nosZ ratio in relation to the ability of the soil to produce N2O.
To evaluate the abundance of denitrifiers relative to total bacteria, the percentages of denitrification genes in proportion to 16S rRNA were calculated. Such calculation was possible due to (i) the similar PCR efficiencies for the different assays and (ii) the use of the same amount of DNA from B. japonicum as a control for the 16S rRNA, nirK, and nosZ assays. Analysis of the percentages of nirK and nosZ in relation to 16S rRNA genes showed very small amounts, between 0.1 and 6%, for both genes (Table 3). Assuming that the 16S rRNA copy number in bacteria varies from 1 to 13 (6), these percentages are in agreement with previous results from cultivation studies showing that the densities of denitrifiers were 2 orders of magnitude lower than those of the total bacteria (2). Similarly, Tiedje (29) reported a denitrifier percentage of the total population ranging between 0.1 and 5%. In contrast, higher ratios of narG to 16S rRNA, up to 40%, were observed. While the 16S rRNA, nirK, and nosZ primers were designed to be as universal as possible, the narG primers used in this study were designed to be specific to a new group of nitrate reducers (15). Even though the narG copy number in this new group is unknown, it does not exceed three copies in known genomes (19). These high percentages of this unknown group of nitrate reducers are in accordance with other reports of their numerical importance in the soil (15). However, from an ecological perspective it is interesting that this novel narG group was less abundant in the three samples from the Himalayas than in all the other soils tested in this study.
In conclusion, the quantitative nosZ PCR assay, which was developed in this study, showed that the N2O-reducing community represented less than 5% of the total bacteria in the studied soils. Since our understanding of microbial communities should be not only qualitative but also quantitative, there is a need in microbial ecology to quantify these microbial populations using cultivation-independent methods to link the diversity and density of the microbial community with its activity (21). The results presented in this study confirm that quantitative PCR, which is specific, sensitive, and rapid, can be used in microbial ecology to quantify genes in large numbers of environmental samples.
Acknowledgments
We thank Olivier Stoiche for helpful technical assistance. We also gratefully acknowledge S. Hallin for providing denitrifying reference strains. Finally, we thank the SSG for providing sequencing facilities.
We thank COST856 “Ecological Aspects of Denitrification, with Emphasis on Agriculture” (http://www.cost856.de) for funding the short-term scientific mission of B. Stres in Dijon. This work was supported by the French Ministry of Research (ACI PNBC “MUTEN”) and by the Burgundy Region.
REFERENCES
- 1.Cavigelli, M. A., and G. P. Robertson. 2001. Role of denitrifier diversity in rates of nitrous oxide consumption in a terrestrial ecosystem. Soil Biol. Biochem. 33:297-310. [Google Scholar]
- 2.Chèneby, D., S. Perrez, C. Devroe, S. Hallet, Y. Couton, F. Bizouard, G. Iuretig, J. C. Germon, and L. Philippot. 2004. Denitrifying bacteria in bulk and maize-rhizospheric soil: diversity and N2O-reducing abilities. Can. J. Microbiol. 50:469-474. [DOI] [PubMed] [Google Scholar]
- 3.Clays-Josserand, A., P. Lemanceau, L. Philippot, and R. Lensi. 1995. Influence of two plant species (flax and tomato) on the distribution of nitrogen dissimilative abilities within fluorescent Pseudomonas spp. Appl. Environ. Microbiol. 61:1745-1749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Coyle, C. L., W. G. Zumft, P. M. Kroneck, H. Korner, and W. Jakob. 1985. Nitrous oxide reductase from denitrifying Pseudomonas perfectomarina. Purification and properties of a novel multicopper enzyme. Eur. J. Biochem. 16:459-467. [DOI] [PubMed] [Google Scholar]
- 5.Coyne, M. S., A. Arunakumari, B. A. Averill, and J. M. Tiedje. 1989. Immunological identification and distribution of dissimilatory cd1 and nonheme copper nitrite reductases in denitrifying bacteria. Appl. Environ. Microbiol. 55:2924-2931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fogel, G. B., C. R. Collins, J. Li, and C. F. Brunk. 1999. Prokaryotic genome size and SSU rDNA copy number: estimation of microbial relative abundance from a mixed population. Microb. Ecol. 38:93-113. [DOI] [PubMed] [Google Scholar]
- 7.Greenberg, E., and G. Becker. 1997. Nitrous oxide as end denitrification product by strains of fluorescent pseudomonads. Can. J. Microbiol. 23:903-907. [DOI] [PubMed] [Google Scholar]
- 8.Henry, S., E. Baudouin, J. C. Lopez-Gutierrez, F. Martin-Laurent, A. Brauman, and L. Philippot. 2004. Quantification of denitrifying bacteria in soils by nirK gene targeted real-time PCR. J. Microbiol. Methods 59:327-335. (Erratum, 61:289-290, 2005.) [DOI] [PubMed] [Google Scholar]
- 9.Holtan-Hartwig, L., P. Dörsch, and L. R. Bakken. 2000. Comparison of denitrifying communities in organic soils: kinetics of NO3− and N2O reduction. Soil Biol. Biochem. 32:833-843. [Google Scholar]
- 10.Horn, M. A., H. L. Drake, and A. Schramm. 2006. Nitrous oxide reductase genes (nosZ) of denitrifying microbial populations in soil and the earthworm gut are phylogenetically similar. Appl. Environ. Microbiol. 72:1019-1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hulse, C. I., and B. A. Averill. 1989. Isolation of a high specific activity pink, monomeric nitrous oxide reductase from Achromobacter cycloclastes. Biophys. Res. Commun. 166:195-199. [DOI] [PubMed] [Google Scholar]
- 12.IPCC. Climate change, the scientific basis. Cambridge University Press, Cambridge, United Kingdom.
- 13.Kloos, K., A. Mergel, C. Rösch, and H. Bothe. 2001. Denitrification within the genus Azospirillum and other associative bacteria. Aust. J. Plant Physiol. 28:991-998. [Google Scholar]
- 14.Kolb, S., C. Knief, S. Stubner, and R. Conrad. 2003. Quantitative detection of methanotrophs in soil by novel pmoA-targeted real-time PCR assays. Appl. Environ. Microbiol. 69:2423-2429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lopez-Gutierrez, J. C., S. Henry, S. Hallet, F. Martin-Laurent, G. Catroux, and L. Philippot. 2004. Quantification of a novel group of nitrate-reducing bacteria in the environment by real-time PCR. J. Microbiol. Methods 57:399-407. [DOI] [PubMed] [Google Scholar]
- 16.Martin-Laurent, F., L. Philippot, S. Hallet, R. Chaussod, J. C. Germon, G. Soulas, and G. Catroux. 2001. DNA extraction from soils: old bias for new microbial diversity analysis methods. Appl. Environ. Microbiol. 67:2354-2359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Michalski, W. P., D. J. Miller, and D. J. D. Nicholas. 1986. Purification and characterization of nitrous oxide reductase from Rhodopseudomonas f.sp. denitrificans. Biochim. Biophys. Acta 872:50-60. [Google Scholar]
- 18.Mounier, E., S. Hallet, D. Chèneby, E. Benizri, Y. Gruet, C. Nguyen, S. Piutti, C. Robin, S. Slezack-Deschaumes, F. Martin-Laurent, J. C. Germon, and L. Philippot. 2004. Influence of maize mucilage on the diversity and activity of the denitrifying community. Environ. Microbiol. 6:301-312. [DOI] [PubMed] [Google Scholar]
- 19.Philippot, L. 2002. Denitrifying genes in bacterial and archeal genomes. Biochim. Biophys. Acta 1577:355-376. [DOI] [PubMed] [Google Scholar]
- 20.Philippot, L. 2006. Use of functional genes to quantify denitrifiers in the environment. Biochem. Soc. Trans. 34:101-103. [DOI] [PubMed] [Google Scholar]
- 21.Philippot, L., and S. Hallin. 2005. Finding the missing link between diversity and activity using denitrifying bacteria as a model functional community. Curr. Opin. Microbiol. 8:234-239. [DOI] [PubMed] [Google Scholar]
- 22.Qiu, X. Y., R. A. Hurt, L. Y. Wu, C. H. Chen, J. M. Tiedje, and J. Z. Zhou. 2004. Detection and quantification of copper-denitrifying bacteria by quantitative competitive PCR. J. Microbiol. Methods 59:199-210. [DOI] [PubMed] [Google Scholar]
- 23.Rich, J. J., and D. D. Myrold. 2004. Community composition and activities of denitrifying bacteria from adjacent agricultural soil, riparian soil, and creek sediment in Oregon, USA. Soil Biol. Biochem. 36:1431-1441. [Google Scholar]
- 24.Rösch, C., A. Mergel, and H. Bothe. 2002. Biodiversity of denitrifying and dinitrogen-fixing bacteria in an acid forest soil. Appl. Environ. Microbiol. 68:3818-3829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Simon, J., O. Einsle, P. M. Kroneck, and W. G. Zumft. 2004. The unprecedented nos gene cluster of Wolinella succinogenes encodes a novel respiratory electron transfer pathway to cytochrome c nitrous oxide reductase. FEBS Lett. 569:7-12. [DOI] [PubMed] [Google Scholar]
- 26.Smith, C. J., D. B. Nedwell, L. F. Dong, and A. M. Osborn. 2006. Evaluation of quantitative polymerase chain reaction-based approaches for determining gene copy and gene transcript numbers in environmental samples. Environ. Microbiol. 8:804-815. [DOI] [PubMed] [Google Scholar]
- 27.Snyder, S. W., and T. C. Hollocher. 1987. Purification and some characteristics of nitrous oxide reductase from Paracoccus denitrificans. J. Biol. Chem. 262:6515-6525. [PubMed] [Google Scholar]
- 28.Stres, B., I. Mahne, G. Avguštin, and J. M. Tiedje. 2004. Nitrous oxide reductase (nosZ) gene fragments differ between native and cultivated Michigan soils. Appl. Environ. Microbiol. 70:301-309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tiedje, J. M. 1988. Ecology of denitrification and dissimilatory nitrate reduction to ammonium, p. 179-244. In A. J. B. Zehnder (ed.), Biology of anaerobic microorganisms. John Wiley & Sons, Inc., New York, N.Y.
- 30.Wood, D. W., J. C. Setubal, R. Kaul, D. Monks, L. Chen, G. E. Wood, Y. Chen, L. Woo, J. P. Kitajima, V. K. Okura, N. F. Almeida, Y. Zhou, D. Bovee, P. Chapman, J. Clendenning, G. Deatherage, W. Gillet, C. Grant, D. Guenthner, T. Kutyavin, R. Levy, M. Li, E. McClelland, A. Palmieri, C. Raymond, G. Rouse, C. Saenphimmachak, Z. Wu, D. Gordon, J. A. Eisen, I. Paulsen, P. Karp, P. Romero, S. Zhang, H. Yoo, Y. Tao, P. Biddle, M. Jung, W. Krespan, M. Perry, B. Gordon-Kamm, L. Liao, S. Kim, C. Hendrick, Z. Zhao, M. Dolan, S. V. Tingey, J. Tomb, M. P. Gordon, M. V. Olson, and E. W. Nester. 2001. The genome of the natural genetic engineer Agrobacterium tumefaciens C58. Science 294:2317-2323. [DOI] [PubMed] [Google Scholar]
- 31.Wrage, N., J. Lauf, A. D. Prado, M. Pinto, S. Pietrzak, S. Yamulki, O. Oenema, and G. Gebauer. 2004. Distinguishing sources of N2O in European grasslands by stable isotope analysis. Rapid Commun. Mass Spectrom. 18:1201-1207. [DOI] [PubMed] [Google Scholar]
- 32.Zumft, G. 1997. Cell biology and molecular basis of denitrification. Microbiol. Mol. Biol. Rev. 61:533-536. [DOI] [PMC free article] [PubMed] [Google Scholar]