Abstract
Transcriptome analyses of Salmonella enterica serovar Typhimurium revealed that 15 genes were significantly up-regulated after 2 h of adaptation with lactic acid. cadB was the most highly up-regulated gene and was shown to be an essential component. Lactic acid-adapted cells exhibited sensitivity to hydrogen peroxide, likely due to down-regulation of the OxyR regulon.
Salmonella enterica serovar Typhimurium employs both a logarithmic-phase acid tolerance response (ATR) and a stationary-phase ATR to cope with acid stress (23). The logarithmic-phase HCl-induced ATR requires both pre- and post-acid-shock adaptation phases to be fully tolerant to a pHo of ≤3.0 at 37°C (13). The pre-acid-shock adaptation phase occurs after exposure to pH 5.8 and involves the production of at least 12 acid shock proteins (5). The gene products of the cad operon (1) and the atp genes (6, 7) all appear to be important in this preshock adaptation phase. Salmonella commonly encounters stress from organic acids, either as by-products of its own metabolism (25), as food preservatives, or as volatile fatty acids in the intestine (12). Limited studies have shown that these organic acids protect against subsequent HCl stress (e.g., gastric acid) (9, 10).
We have recently shown that lactic acid induces a transient ATR in Salmonella serovar Typhimurium at 20°C, and the maximum ATR occurs at 2 h (9). We used a transcriptome approach to identify the genes involved in the lactic acid-induced ATR (L-ATR). Mid-logarithmic cultures of Salmonella serovar Typhimurium SL1344 were produced and adapted for 2 h with 0.015 M lactic acid in tryptone soy broth (TSB) (Difco) at pHo 5.8 as previously described (9). After adaptation, RNA was stabilized by addition of a phenol-ethanol solution (5:95) and was immediately transferred to ice prior to extraction. RNA samples were prepared using a Promega SV total RNA purification kit, and the quality was checked using an RNA nanochip (with an Agilent 2100 bioanalyzer). The microarray experiment and analyses of transcriptome data were performed as previously described (4). We used an indirect “type 2” experimental design (26) with genomic DNA as a common reference. We used two technical replicates for each of two biological replicates. The relevant protocols are described at http://www.ifr.bbsrc.ac.uk/safety/microarrays/protocols.html. The 4,114-feature microarray based on the serovar Typhimurium LT2 genome sequence (18) has been described previously (4).
Following statistical filtering (false discovery rate, 0.05), changes in gene expression of >twofold were considered to be significant (4). A total of 76 genes changed during the L-ATR, and 15 of them were significantly up-regulated (see Table S1 in the supplemental material). Only three of these up-regulated genes, cadA, cadB, and cspB, exhibited more than a fivefold increase in the level of expression (Table 1). Expression of cadBA can be induced in Escherichia coli by lowering the external pH in the presence of lysine (20). This raises the possibility that the high level of cadBA induction observed in the L-ATR may not have been caused by lactic acid per se but could have reflected the decrease in the pHo in the presence of lysine in the medium.
TABLE 1.
Salmonella serovar Typhimurium genes exhibiting more than a fivefold increase in the level of expression during the lactic acid-induced acid tolerance response and members of the OxyR regulon that are down-regulated during the responsea
| Gene | Expression level after exposure to lactic acid at pH 5.8 compared to expression at pH 7 | Function or comment | |
|---|---|---|---|
| Common name | Systematic name | ||
| cadA | STM2559 | 5.9 | Lysine decarboxylase 1 |
| cadB | STM2558 | 50.94 | Lysine/cadaverine transport protein |
| cspB | STM1996 | 8.97 | Putative cold shock protein |
| ahpC | STM0608 | 0.2 | Detoxification of hydroperoxides |
| ahpF | STM0609 | 0.06 | Detoxification of hydroperoxides |
| dps | STM0831 | 0.14 | Starvation-induced resistance to H2O2 |
| katG | STM4106 | 0.09 | Catalase; hydroperoxidase HPI(I) |
| oxyR | STM4125 | 0.57 | Regulatory protein sensor for oxidative stress |
The complete set of transcriptome data has been submitted to the ArrayExpress database (www.ebi.ac.uk/arrayexpress).
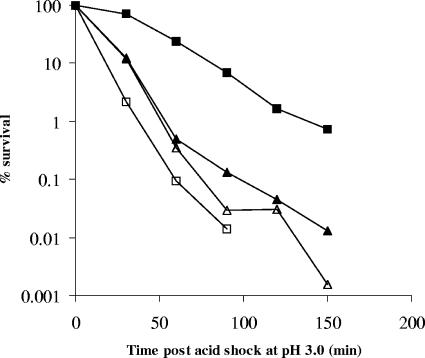
The cadB gene is a component of the lysine decarboxylase system and has a known role in the HCl-induced ATR of Salmonella serovar Typhimurium (21). As cadB was the most highly up-regulated gene in the L-ATR, its role was investigated further. Using the λ Red method (3), we replaced the cadB structural gene in SL1344 with a kanamycin resistance marker to generate strain JH2917. The mutation was P22 transduced to a wild-type SL1344 background, and the genotype of the mutant strain was verified by PCR. The following primers were used for PCR amplification of the DNA fragment that was recombined with the chromosome from the pKD4 plasmid (3): CadB-KO-F (5′-TAAGCCCGGTTCTTAAAAATACAGCTCAGGAGAAATGAACGTGTAGGCTGGAGCTGCTTC) and CadB-KO-R (5′-GCAGGTGACGAAAGGGGCTTTGAGAAAAAGGAGTTAGCGGCATATGAATATCCTCCTTA). It is possible that the cadB::kan mutation could have a polar effect on the downstream cadA gene. Even if this were the case, the mutant phenotype would remain as a defect in lysine decarboxylase. To test whether the lysine decarboxylase system was required for the L-ATR, cultures of Salmonella serovar Typhimurium SL1344 and JH2917 were grown to the mid-logarithmic phase and either adapted with lactic acid prior to acid shock or immediately acid shocked (unadapted cultures) as described previously (9). JH2917 was not able to mount an ATR (Fig. 1), showing that the lysine decarboxylase system was an essential component of the L-ATR. CadA has been shown to be important in the ATR of Vibrio cholerae (19), and CadB has been shown to be important but not essential for the HCl-induced ATR in Salmonella serovar Typhimurium (8, 21), in which other amino acid decarboxylase systems can be invoked to restore the ATR phenotype (21). However, our study demonstrated that JH2917 was unable to mount an ATR even though the organism was provided with a rich source of alternate amino acids. Recently, it has been shown that the inhibition of porins by excreted cadaverine may be part of the adaptive mechanism that helps E. coli survive in acidic environments (24). Our results indicate that a similar adaptive mechanism may be essential for the Salmonella serovar Typhimurium L-ATR, as it requires cadaverine transport mediated by the CadB transport protein.
FIG. 1.
Survival of lactic acid-adapted ΔcadB mutant JH2917 and wild-type Salmonella serovar Typhimurium strain SL1344 during acid shock at an external pH of 3.0 poised with HCl. Cells were adapted at pHo 5.8 for 2 h. □, unadapted wild-type cells; ▪, adapted wild-type cells; ▵, unadapted JH2917 cells; ▴, adapted JH2917 cells. The values are means for two biological replicates. The coefficient of variation was <20%.
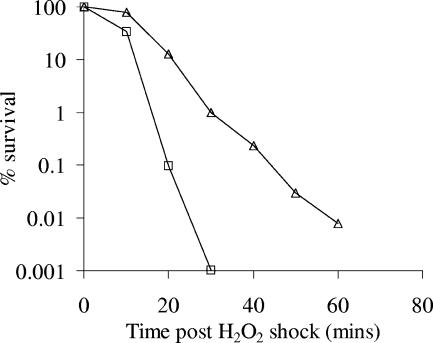
The transcriptome data also showed that five members of the OxyR regulon were down-regulated during the L-ATR (Table 1); this regulon is critical for the survival of Salmonella under oxidative stress conditions (2). Such down-regulation during the L-ATR raised the possibility that although adaptation with lactic acid protected against acidic environments, it was incompatible with the response to oxidative stress. To investigate this, lactic acid-adapted and unadapted cultures of Salmonella serovar Typhimurium SL1344 were exposed to an oxidative stress. Cells were removed from TSB by filtration through a 0.22-μm membrane filter (Millipore) and were resuspended by vortexing in fresh TSB, and enough of a 30% (vol/vol) solution of hydrogen peroxide (H2O2) (Sigma) was added to obtain a concentration of 100 mM. Cultures were then reincubated at 20°C at 120 rpm, and the number of viable cells was determined, as previously described (9), every 10 min for 1 h. As predicted, the L-ATR cells were vulnerable to oxidative damage and displayed a hypersensitive phenotype compared with unadapted cells (Fig. 2). Cross-protection between different stressors has been well documented (11, 15, 17, 22). In Salmonella spp., acid adaptation has been shown to result in tolerance to H2O2, heat, salt, crystal violet, and polymyxin B (11, 14, 15). However, acid adaptation has also been shown to sensitize Salmonella serovar Typhimurium to hypochlorous acid (16). Our study demonstrated that the L-ATR in Salmonella serovar Typhimurium causes sensitivity to hydrogen peroxide; the L-ATR appears to inactivate the oxidative stress response via down-regulation of the OxyR regulon. Therefore, cells exhibiting the L-ATR are protected against acidic environments but are vulnerable to oxidative stress, a phenotype that could be exploited in preservation regimens.
FIG. 2.
Survival of lactic acid-adapted and unadapted cells of Salmonella serovar Typhimurium SL1344 during exposure to hydrogen peroxide. Cells were adapted with lactic acid at pHo 5.8 for 2 h. The bacterial cells were subsequently challenged with 100 mM H2O2. ▵, unadapted cells; □, lactic acid-adapted cells. The values are means for two biological replicates. The coefficient of variation was <20%.
This L-ATR-linked peroxide sensitivity contrasts with findings of other studies in which ATR-linked resistance to hydrogen peroxide was identified (11, 14). In both previous studies the workers found that adaptation of Salmonella serovar Typhimurium with HCl (14) or mixtures of short-chain fatty acids (11) induced resistance to hydrogen peroxide, and it is possible that the opposing hydrogen peroxide phenotypes were a consequence of the type of acidulants used in the experiments.
In this study, we identified genes involved in the L-ATR of Salmonella serovar Typhimurium and related transcriptional changes in bacterial phenotypes, including resistance to acidity and sensitivity to oxidative stress. This type of approach highlights the value of transcriptome analysis with DNA microarrays for the food industry, since the protective responses and any linked vulnerabilities associated with particular environmental stresses can be identified and exploited to enhance food preservation regimens.
Supplementary Material
Acknowledgments
We acknowledge financial support from a core strategic grant from the UK Biotechnology and Biological Sciences Research Council for this work.
We are grateful to John Foster and David Wilson for helpful discussions and to Matt Rolfe for microarray construction.
Footnotes
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Bearson, S., B. Bearson, and J. W. Foster. 1997. Acid stress responses in enterobacteria. FEMS Microbiol. Lett. 147:173-180. [DOI] [PubMed] [Google Scholar]
- 2.Christman, M. F., R. W. Morgan, F. S. Jacobson, and B. N. Ames. 1985. Positive control of a regulon for defenses against oxidative stress and some heat-shock proteins in Salmonella typhimurium. Cell 41:753-762. [DOI] [PubMed] [Google Scholar]
- 3.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 97:6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Eriksson, S., S. Lucchini, A. Thompson, M. Rhen, and J. C. D. Hinton. 2003. Unravelling the biology of macrophage infection by gene expression profiling of intracellular Salmonella enterica. Mol. Microbiol. 47:103-118. [DOI] [PubMed] [Google Scholar]
- 5.Foster, J. W., and H. K. Hall. 1990. Adaptive acidification tolerance response of Salmonella typhimurium. J. Bacteriol. 172:771-778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Foster, J. W., and H. K. Hall. 1991. Inducible pH homeostasis and the acid tolerance response of Salmonella typhimurium. J. Bacteriol. 173:5129-5135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Foster, J. W. 1993. The acid tolerance response of Salmonella typhimurium involves transient synthesis of key acid shock proteins. J. Bacteriol. 175:1981-1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Foster, J. W., Y. K. Park, I. S. Bang, K. Karem, H. Betts, H. K. Hall, and E. Shaw. 1994. Regulatory circuits involved with pH-regulated gene expression in Salmonella typhimurium. Microbiology 140:341-352. [DOI] [PubMed] [Google Scholar]
- 9.Greenacre, E. J., T. F. Brocklehurst, C. R. Waspe, D. R. Wilson, and P. D. G. Wilson. 2003. Salmonella enterica serovar Typhimurium and Listeria monocytogenes acid tolerance response induced by organic acids at 20°C: optimization and modeling. Appl. Environ. Microbiol. 69:3945-3951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kwon, Y. M., and S. C. Ricke. 1998. Induction of acid resistance of Salmonella typhimurium by exposure to short-chain fatty acids. Appl. Environ. Microbiol. 64:3458-3463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kwon, Y. M., S. Y. Park, S. G. Birkhold, and S. C. Ricke. 2000. Induction of resistance of Salmonella typhimurium to environmental stresses by exposure to short-chain fatty acids. J. Food Sci. 65:1037-1040. [Google Scholar]
- 12.Lawhon, S. D., R. Maurer, M. Suyemoto, and C. Altier. 2002. Intestinal short-chain fatty acids alter Salmonella typhimurium invasion gene expression and virulence through BarA/SirA. Mol. Microbiol. 46:1451-1464. [DOI] [PubMed] [Google Scholar]
- 13.Lee, I. S., J. L. Slonczewski, and J. W. Foster. 1994. A low-pH-inducible, stationary-phase acid tolerance response in Salmonella typhimurium. J. Bacteriol. 176:1422-1426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee, I. S., J. S. Lin, H. K. Hall, B. Bearson, and J. W. Foster. 1995. The stationary-phase sigma-factor sigma(S) (Rpos) is required for a sustained acid tolerance response in virulent Salmonella typhimurium. Mol. Microbiol. 17:155-167. [DOI] [PubMed] [Google Scholar]
- 15.Leyer, G. J., and E. A. Johnson. 1993. Acid adaptation induces cross-protection against environmental stresses in Salmonella typhimurium. Appl. Environ. Microbiol. 59:1842-1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Leyer, G. J., and E. A. Johnson. 1997. Acid adaptation sensitizes Salmonella typhimurium to hypochlorous acid. Appl. Environ. Microbiol. 63:461-467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mattick, K. L., F. Jorgensen, J. D. Legan, H. M. Lappin-Scott, and T. J. Humphrey. 2000. Habituation of Salmonella spp. at reduced water activity and its effect on heat tolerance. Appl. Environ. Microbiol. 4921-4925. [DOI] [PMC free article] [PubMed]
- 18.McClelland, M., K. E. Sanderson, J. Spieth, S. W. Clifton, P. Latreille, L. Courtney, S. Porwollik, J. Ali, M. Dante, F. Y. Du, S. F. Hou, D. Layman, S. Leonard, C. Nguyen, K. Scott, A. Holmes, N. Grewal, E. Mulvaney, E. Ryan, H. Sun, L. Florea, W. Miller, T. Stoneking, M. Nhan, R. Waterston, and R. K. Wilson. 2001. Complete genome sequence of Salmonella enterica serovar Typhimurium LT2. Nature 413:852-856. [DOI] [PubMed] [Google Scholar]
- 19.Merrell, D. S., and A. Camilli. 1999. The cadA gene of Vibrio cholerae is induced during infection and plays a role in acid tolerance. Mol. Microbiol. 34:836-849. [DOI] [PubMed] [Google Scholar]
- 20.Neely, M. N., and E. R. Olson. 1996. Kinetics of expression of the Escherichia coli cad operon as a function of pH and lysine. J. Bacteriol. 178:5522-5528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Park, Y. K., B. Bearson, S. H. Bang, I. S. Bang, and J. W. Foster. 1996. Internal pH crisis, lysine decarboxylase and the acid tolerance response of Salmonella typhimurium. Mol. Microbiol. 20:605-611. [DOI] [PubMed] [Google Scholar]
- 22.Rowe, M. T., and R. Kirk. 1999. An investigation into the phenomenon of cross-protection in Escherichia coli O157:H7. Food Microbiol. 16:157-164. [Google Scholar]
- 23.Rychlik, I., and P. A. Barrow. 2005. Salmonella stress management and its relevance to behaviour during intestinal colonisation and infection. FEMS Microbiol. Rev. 29:1021-1040. [DOI] [PubMed] [Google Scholar]
- 24.Samartzidou, H., M. Mehrazin, Z. H. Xu, M. J. Benedik, and A. H. Delcour. 2003. Cadaverine inhibition of porin plays a role in cell survival at acidic pH. J. Bacteriol. 185:13-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wilson, P. D. G., D. R. Wilson, T. F. Brocklehurst, H. P. Coleman, G. Mitchell, C. R. Waspe, S. A. Jukes, and M. M. Robins. 2003. Batch growth of Salmonella typhimurium LT2: stoichiometry and factors leading to cessation of growth. Int. J. Food Microbiol. 89:195-203. [DOI] [PubMed] [Google Scholar]
- 26.Yang, Y. H., and T. Speed. 2002. Design issues for cDNA microarray experiments. Nat. Rev. Genet. 3:579-588. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.