Abstract
A bacterium designated Pseudonocardia sp. strain ENV478 was isolated by enrichment culturing on tetrahydrofuran (THF) and was screened to determine its ability to degrade a range of ether pollutants. After growth on THF, strain ENV478 degraded THF (63 mg/h/g total suspended solids [TSS]), 1,4-dioxane (21 mg/h/g TSS), 1,3-dioxolane (19 mg/h/g TSS), bis-2-chloroethylether (BCEE) (12 mg/h/g TSS), and methyl tert-butyl ether (MTBE) (9.1 mg/h/g TSS). Although the highest rates of 1,4-dioxane degradation occurred after growth on THF, strain ENV478 also degraded 1,4-dioxane after growth on sucrose, lactate, yeast extract, 2-propanol, and propane, indicating that there was some level of constitutive degradative activity. The BCEE degradation rates were about threefold higher after growth on propane (32 mg/h/g TSS) than after growth on THF, and MTBE degradation resulted in accumulation of tert-butyl alcohol. Degradation of 1,4-dioxane resulted in accumulation of 2-hydroxyethoxyacetic acid (2HEAA). Despite its inability to grow on 1,4-dioxane, strain ENV478 degraded this compound for >80 days in aquifer microcosms. Our results suggest that the inability of strain ENV478 and possibly other THF-degrading bacteria to grow on 1,4-dioxane is related to their inability to efficiently metabolize the 1,4-dioxane degradation product 2HEAA but that strain ENV478 may nonetheless be useful as a biocatalyst for remediating 1,4-dioxane-contaminated aquifers.
Ether-containing organic compounds are widely used as solvents, pesticides, and gasoline additives and in a host of other applications. Very recently, the solvent stabilizer 1,4-dioxane has emerged as an important groundwater contaminant throughout the United States and elsewhere (26). Like many ethers, this compound is miscible in water, has a low dimensionless Henry's Law constant (2.0 × 10−4), and has a low octanol/water partitioning coefficient (1.23); thus, it is poorly retarded in aquifers and has the potential to create large contaminant plumes that threaten drinking water supplies that are distant from the original release sites (14, 23, 29).
Few treatment methods have proven to be successful and economically feasible for removing 1,4-dioxane from groundwater. Because of the low octanol/water partitioning coefficient and low Henry's Law constant of this compound, traditional remediation technologies like carbon adsorption and air stripping are inefficient and costly. Likewise, in situ and ex situ biological treatments of 1,4-dioxane have not emerged as viable treatment options even though some microbes have been shown to degrade the compound (3, 5, 12, 18, 23, 27, 28, 43). Only a few ex situ technologies, including chemical oxidation with a combination of ozone and hydrogen peroxide (1) or hydrogen peroxide and UV light (32), have been utilized commercially to destroy 1,4-dioxane, but the cost of using these technologies for high-concentration waste streams can be prohibitive.
Relatively few studies have evaluated biological degradation of 1,4-dioxane, but the indigenous microorganisms at contaminated sites often are not able to degrade this compound (12, 23). In the last several years, however, 1,4-dioxane biodegradation has been reported for both pure (28, 5, 3) and mixed cultures of bacteria (18, 43) and for a fungal isolate (27). For example, a pure culture of the propane-oxidizing bacterium Mycobacterium vaccae JOB5 was shown to partially degrade 1,4-dioxane but not to grow on this compound (5). Bernhardt and Diekmann (3) reported biodegradation of 1,4-dioxane by a Rhodococcus strain, and Parales et al. (28) isolated a bacterium (strain CB1190) that is capable of sustained growth and mineralization of 1,4-dioxane, albeit at low rates. Strain CB1190 has recently been reclassified as Pseudonocardia dioxanivorans strain CB1190 (24). In other studies, mixed cultures of bacteria were able to degrade 1,4-dioxane, but only in the presence of the cosubstrate tetrahydrofuran (THF) (43).
Unlike biodegradation of 1,4-dioxane, biodegradation of the cyclic monoether THF has been well studied, and this compound appears to be a growth substrate for many bacteria (3, 7, 19, 28, 37, 43). Detailed molecular and biochemical analysis of THF degradation by Pseudonocardia sp. strain K1 led to cloning of an operon involved in THF degradation and to description of a biodegradation pathway for THF (37). The initial transformation of THF by strain K1 appears to involve a binuclear iron-containing multicomponent THF monooxygenase (THFmo) that oxidizes THF to 2-hydroxytetrahydrofuran (36, 37). The authors also suggested that a dehydrogenase could convert 2-hydroxytetrahydrofuran to γ-butyrolactone, a hydrolase could convert γ-butyrolactone to 4-hydroxybutyrate, and a second dehydrogenase could convert 4-hydroxybutyrate to succinate semialdehyde.
Although no bacterial 1,4-dioxane biodegradation pathway has been described, two reports have described 1,4-dioxane degradation in rats. Woo et al. (40) suggested that 1,4-dioxane-2-one (PDX) is the major urinary metabolite of 1,4-dioxane in rats but that there could be other pathways. Braun and Young (4), using a different approach, identified 2-hydroxyethoxyacetic acid (2HEAA) as the major metabolite. δ-Hydroxy acids, however, rarely are present in the pure state or in aqueous solutions except in the form of salts; rather, the majority of these acids are known primarily in the lactone form. Thus, lactonization of 2HEAA would result in production of PDX, as identified in the study of Woo et al. (40). PDX also can polymerize spontaneously to form linear polymers, or it can be reversibly converted to 2HEAA under basic conditions. In a study of 1,4-dioxane degradation by a pure culture of filamentous fungi, ethylene glycol was the first 1,4-dioxane product detected, suggesting that there may be an alternate degradation pathway (27).
In this paper we describe isolation and characterization of a new THF-degrading bacterium, strain ENV478. We evaluated the ability of this strain to degrade ether-containing pollutants and focused on 1,4-dioxane degradation. The initial 1,4-dioxane biodegradation experiments revealed that 2HEAA is produced as an apparent terminal product of 1,4-dioxane degradation in strain ENV478. These results suggest that the inability of strain ENV478 and perhaps other THF-degrading strains to grow on 1,4-dioxane may be related to their inability to efficiently utilize this metabolite.
MATERIALS AND METHODS
Chemicals.
1,4-Dioxane (98%) was purchased from Aldrich Chemical Co. (Milwaukee, WI). R2A medium was obtained from BBL, Inc. (Cockeysville, MD). Unless indicated otherwise, all other chemicals were the highest purity available and were purchased from either Aldrich Chemical Co. (Milwaukee, WI), Mallinckrodt Specialty Chemical Co. (Paris, KY), J. T. Baker Inc. (Phillipsburg, NJ), or Sigma Chemical Co. (St. Louis, MO). Uniformly labeled 1,4-[14C]dioxane (0.08 mCi/mmol; 96.4% radiochemical purity as determined by high-performance liquid chromatography [HPLC]) was purchased from Sigma (St. Louis, MO).
Isolation and growth of strain ENV478.
Strain ENV478 was isolated by enrichment culturing from the liquor of a membrane bioreactor system that was treating a mixture of wastes from a fine chemical manufacturer. Enrichment culturing was performed with THF (100 mg/liter) as the sole carbon source and basal salts medium (BSM) (13). Cultures growing on THF were plated on R2A agar plates (EM Science, Gibbstown, NJ), and individual colonies were screened for the ability to grow on THF as a sole source of carbon and energy. Individual isolates were grown in BSM containing THF, washed, and screened as described below for the ability to degrade 1,4-dioxane. Strain ENV478 was maintained on R2A agar plates or in BSM containing THF. Enrichment culturing of the same reactor liquor with 1,4-dioxane as the sole carbon source did not result in a culture that could grow on 1,4-dioxane.
16S rRNA analysis.
Total genomic DNA was isolated from strain ENV478 by using a DNA extraction kit (Clontech, Mountain View, CA). The 16S rRNA genes were PCR amplified with primers 27f and 1522r under conditions recommended by the supplier (Sigma, St. Louis, MO). The purified PCR product (QIAGEN, Valencia, CA) was used directly in DNA sequencing reactions (Applied Biosystems, Foster City, CA) with primers 27f, 357f, 704f, 926f, 1242f, 342r, 685r, 907r, 1392r, and 1522r (15). The complete sequence was assembled and edited with the Lasergene program (DNAStar, Madison, WI). Closely matching sequences were found in the GenBank database using the BLAST algorithm (2), as well as the search function of the Ribosomal Database Project (6). Sequences were aligned using CLUSTALV in the Lasergene software package (DNAStar) and were visually inspected. Phylogenetic analysis was performed with the MEGA version 2.1 software package (20) Distances were determined by maximum parsimony, and bootstrap values were calculated by using 1,000 replications.
Biodegradation assays.
Unless indicated otherwise, the cells of strain ENV478 used in biodegradation assays were grown on THF in BSM at 30°C. The cells were collected by centrifugation, washed twice in BSM, and suspended in fresh BSM. Because of the tendency of the cells to clump, cell density was typically determined by measuring total suspended solids (TSS) (39), a measurement of the dry weight of filtered cells. Assays were performed in 60-ml serum vials sealed with Teflon-lined septa and aluminum crimp seals. Killed control samples were poisoned with HgCl2 (final concentration, 0.1 mM). Ethers were analyzed by using gas chromatography-mass spectrometry (GC-MS) (38) with heated purge and trap.
Synthesis of potential pathway intermediates.
Elucidation of the pathway required that we first synthesize the potential degradation products as analytical standards. 2HEAA and PDX were synthesized essentially as described by Doddi et al. (9). This synthesis involved preparation of sodium glycoxide by reacting excess ethylene glycol with metallic sodium. The glycoxide was then reacted with chloroacetic acid to obtain the sodium salt of 2HEAA (NaHEA). The synthesis reactions are described by the following equations:
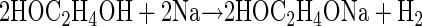 |
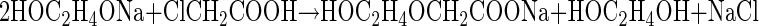 |
Excess ethylene glycol and reaction by-products were removed by distillation and by washing the residual solid with acetone. The NaHEA was then purified by hot filtering and recrystallization in ethanol-10% water. 2HEAA was prepared in an aqueous solution by eluting a solution of NaHEA through a cation-exchange resin column in the H+ form.
PDX was prepared by acidifying NaHEA in ethanol with hydrochloric acid to make 2HEAA and filtering the preparation to remove NaCl. The filtrate containing 2HEAA was distilled at atmospheric pressure over MgCO3 to remove ethanol and then slowly heated to distill off product, and the fraction boiling at approximately 200°C was collected.
The sodium glycolate and chloroacetic acid synthesis procedure produced NaHEA with a yield of purified product of approximately 60%. The identity of the product was verified by GC-MS analysis of the methyl ester derivative of NaHEA, using the method described by Braun and Young (4). This produced a characteristic [M-18] response peak at m/e 116 and additional peaks at m/e 75, 87, and 103.
The PDX preparation procedure produced a viscous colorless liquid that slowly solidified to a wax after standing. The identity of PDX was verified by direct GC-MS analysis in methylene chloride extracts. The GC-MS chromatogram peak produced a primary [M] response at m/e 102 and additional peaks at m/e 58 and 73.
Identification of 1,4-[14C]dioxane degradation products.
Ten-milliliter subsamples of a THF-grown ENV478 culture were added to 60-ml serum vials that were sealed with Teflon-lined septa. Ten microliters of a 50:50 mixture of methanol and [14C]dioxane (0.47 μCi) was injected into the vials along with 2 μl of neat THF. The THF was included to ensure that all of the THF degradation genes were induced. The disappearance of 1,4-dioxane was monitored by injecting 1 μl of a sample into a Varian 3400 gas chromatograph (Varian, Walnut Creek, Calif.) equipped with a 30-m capillary Vocol column (Supelco, Inc., Bellefonte, Pa.) and a flame ionization detector. The injector and detector temperatures were maintained at 180 and 220°C, respectively, whereas the column temperature was programmed to increase from 85°C to 140°C at a rate of 50°C/min. When ∼80% of the dioxane had been degraded, an HPLC analysis was performed to monitor the disappearance of 1,4-dioxane and the appearance of biodegradation metabolites. To prepare the samples for analysis, 500 μl of sample was placed in a microcentrifuge tube and centrifuged for 5 min at 14,000 rpm (16,000 × g) to remove the cellular material. The supernatant was then transferred to a 1-ml autosampler vial, which was sealed with a Teflon septum. The metabolites were resolved with a Restek Ultra Aqueous C18 column (3.2 by 150 mm; Restek, Bellefonte, PA) and an isocratic mixture of methanol and water (5:95) at a flow rate of 0.4 ml/min. Elution of the metabolites from the column was monitored at a wavelength of 210 nm. The HPLC eluant was divided into fractions (5- to 30-s intervals) by collecting the eluant in scintillation vials prefilled with 3 ml of scintillation cocktail so that the amount of radioactivity in each fraction and the total radioactivity in the supernatant could be determined. The elution order of the metabolites was determined by spiking authentic standards with radiolabeled products.
Microcosm studies.
Soil and groundwater samples were collected by Solutions-IES (Raleigh, NC) from a 1,4-dioxane-contaminated aquifer located near Elkton, MD. Soil core samples were recovered from the saturated zone of the aquifer from 2.4 to 3.6 m below the surface by using direct push technology. The core sample was cut to fit in a cooler, and the ends of the core sections were sealed. Three liters of groundwater was collected by using a peristaltic pump and stored in sterile glass jars. All samples were placed on ice and shipped via overnight courier to our laboratory. Soil samples were extruded from the cores, combined, and mixed in an anaerobic chamber, and then they were passed through a 6.4-mm sieve to remove stones. Microcosms were constructed in triplicate 160-ml serum vials, and they received 40 g of soil and 80 ml of groundwater. Control microcosms received no amendments. Killed samples received 1 ml of 3.6% HgCl2, and augmented samples received 1 ml of washed THF-grown ENV478 (optical density at 550 nm, 16). The microcosms were sealed with Teflon-lined septa and aluminum crimp seals, and 20 ml of oxygen was injected into each microcosm headspace. The microcosms were placed on their sides and incubated at 15°C on an orbital shaker (120 rpm) in the dark. For sampling, the microcosms were shaken vigorously and the soil was allowed to settle before 1 to 5 ml of the aqueous fraction was removed with a syringe and analyzed by GC-MS as described above. In addition to 1,4-dioxane (∼100 μg/liter), the microcosms contained cis-1,2-dichloroethene (110 μg/liter), 1,1-dichloroethane (74 μg/liter), 1,1,1-trichloroethane (600 μg/liter), and trichloroethene (200 μg/liter), and they had a total organic carbon content of 1.2 mg/liter.
Nucleotide sequence accession number.
The strain ENV478 16S rRNA sequence has been deposited in the GenBank database under accession number DQ437530.
RESULTS
Strain ENV478.
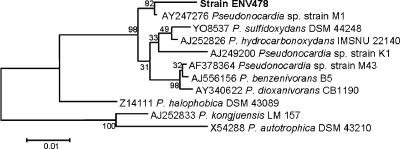
Enrichment culturing of an industrial wastewater treatment system resulted in isolation of several bacterial isolates that could grow on THF as the sole carbon and energy source. None of the organisms isolated, however, were able to grow on 1,4-dioxane. One isolate was selected for further study and was designated strain ENV478. Analysis of the 16S rRNA gene sequence of this strain indicated that it is a member of the genus Pseudonocardia and clusters with Pseudonocardia benzenivorans, Pseudonocardia dioxanivorans, Pseudonocardia hydrocarbonoxydans, and Pseudonocardia sulfidoxydans (Fig. 1). The ability to degrade tetrahydrofuran is well known in this cluster; strains M1, K1, and CB1190 all have this characteristic (7, 19, 24). Given the 16S rRNA sequence distances between P. benzenivorans, P. dioxanivorans, P. hydrocarbonoxydans, and P. sulfidoxydans and the sequence distance between strain ENV478 and these species, strain ENV478 is probably a member of a new Pseudonocardia species.
FIG. 1.
Dendrogram showing the phylogenetic relationship of strain ENV478 to closely related Pseudonocardia strains. The GenBank accession number, genus, species (if known), and strain designation are indicated for each 16S rRNA sequence. The 16S rRNA sequences used for P. autotrophica, P. benzenivorans, P. dioxanivorans, P. halophobica, P. hydrocarbonoxydans, P. kongjuensis, and P. sulfidoxydans are the sequences of the type strains of the species (10, 17, 24, 25, 21, 22, 30). Strains M1, K1, and M43 have not been assigned to species (7, 19). The bootstrap values at the nodes indicate the percentages of occurrence in 1,000 bootstrapped trees. Bar, genetic distance of 0.01.
When grown on BSM with THF, the strain ENV478 culture produced dense clumps of cells that made accurate sampling and measurement of optical density difficult. The culture grew readily on BSM with yeast extract and on R2A agar plates. It also grew on lactate, propane, and sucrose, as well as on the following compounds at a concentration of 200 mg/liter each: 1-propanol, 2-propanol, 1-butanol, diethyl ether, diisopropyl ether, propionic acid, butanoic acid, pentanoic acid, and hexanoic acid. It grew on 50 mg/liter octanoic acid but not on 100 or 200 mg/liter octanoic acid. It did not grow on octanol or hexanol, and growth on 1,4-dioxane and 1,3-dioxolane was slight or nonexistent after 30 days of incubation.
Biodegradation of 1,4-dioxane.
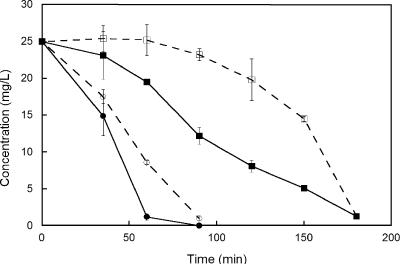
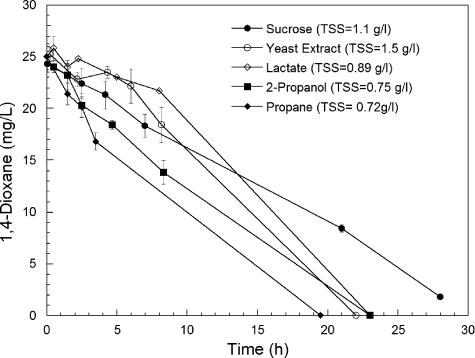
Following growth on THF, strain ENV478 degraded 1,4-dioxane at an initial rate of approximately 21 mg/h/g TSS, which was approximately one-third the rate at which it degraded THF (63 mg/h/g TSS). When the two substrates were added together at equal concentrations, 1,4-dioxane degradation did not occur until the THF was removed from the medium (Fig. 2). When similarly grown cells were incubated with both 1,4-dioxane and 1,3-dioxolane, they degraded both compounds simultaneously and at approximately the same rate (data not shown). Strain ENV478 also degraded 1,4-dioxane after growth on sucrose (0.71 mg/h/g TSS), yeast extract (1.1 mg/h/g TSS), sodium lactate (0.6 mg/h/g TSS), 2-propanol (1.5 mg/h/g TSS), and propane (3.2 mg/h/g TSS) (Fig. 3), but the 1,4-dioxane degradation rates were much lower than those observed with THF-grown cells (21 mg/h/g TSS). After growth on lactate, the strain also degraded 1,3-dioxolane (1.0 mg/h/g TSS). These findings suggested that strain ENV478 has a low level of constitutive activity with these ethers.
FIG. 2.
Biodegradation of 1,4-dioxane (squares) and THF (circles) by strain ENV478 when the compounds were added alone (solid symbols) or as a 50:50 mixture (open symbols). The error bars indicate one standard error of the mean (n = 3).
FIG. 3.
Biodegradation of 1,4-dioxane by strain ENV478 after growth on different substrates. The initial cell densities (TSS) are indicated. The error bars indicate one standard error of the mean (n = 3).
In addition to strain ENV478, for comparison we evaluated 1,4-dioxane degradation by our previously described ether-degrading propanotroph strain ENV425 (34). Strain ENV425 also grew on THF and degraded 1,4-dioxane, but at only one-half the rate (10 mg/h/gTSS) of strain ENV478. Furthermore, whereas strain ENV478 degraded 1,795 mg 1,4-dioxane/g TSS after THF feeding was stopped, strain ENV425 degraded only 200 mg 1,4-dioxane/g TSS.
Analysis of 1,4-dioxane metabolites.
Attempts to isolate chemically synthesized 2HEAA from aqueous solutions after acidification of NaHEA were hampered by apparent conversion of 2HEAA to PDX in a pH-driven equilibrium (4). The equilibrium was driven toward PDX at low pHs in the range from pH 2 to 3, and this was demonstrated by the appearance of PDX in solutions of NaHEA after acidification, as determined by GC-MS analysis of solution extracts. The equilibrium was difficult to monitor because we could not analyze both PDX and HEAA in a single experiment without the potential for interconversion. Direct GC-MS analysis of PDX product solutions revealed only PDX due to our inability to see 2HEAA without derivatization to the methyl ester. Analysis of PDX product solutions for 2HEAA by the methyl ester derivatization method showed that both PDX and 2HEAA were present at a ratio of about 1:4, but the methyl ester 2HEAA derivative may have been formed from PDX during the derivatization process. This made identification and determination of the PDX purity and analysis of PDX in cultures difficult because the derivatization method may result in identification of both compounds as 2HEAA.
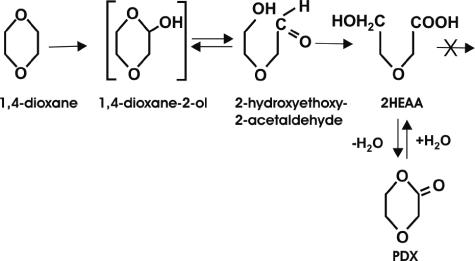
To further evaluate the extent of degradation of 1,4-dioxane by strain ENV478, THF-grown cells were washed and incubated with uniformly labeled 1,4-[14C]dioxane, and an HPLC method that did not require derivatization was developed and employed. Analysis of the culture liquor demonstrated that the [14C]dioxane was converted to a product that coeluted with authentic 2HEAA, but no other potential intermediates (Fig. 4) were detected. The apparent 2HEAA product was not degraded further, even after 402 h of incubation at 25°C. Addition of THF to cultures that had depleted the 1,4-dioxane and accumulated 2HEAA did not result in further degradation of this product. At the end of the experiment, no overall decrease in the radioactivity in the supernatant was observed compared to the radioactivity in the control, indicating that no carbon dioxide had been produced and no 1,4-dioxane was converted to strain ENV478 biomass. In separate growth studies, strain ENV478 was not able to grow on HEAA as a sole carbon source.
FIG. 4.
Proposed partial pathway for biodegradation of 1,4-dioxane by strain ENV478. The 1,4-dioxane-2-ol (hemiacetal) and 2-hydroxyethoxy-2-acetaldehyde were not detectable in the culture media by the methods used in this study. PDX is expected to form spontaneously from 2HEAA under certain conditions (42), but it was not detectable in our experiments.
Degradation of other ethers.
In other degradation experiments with THF-grown strain ENV478, the culture was able to degrade the related solvent 1,3-dioxolane (19 mg/h/g TSS), the gasoline additive methyl tert-butyl ether (MTBE) (9.1 mg/h/g TSS), and the plasticizer bis-2-chloroethyl ether (BCEE) (12 mg/h/g TSS). Interestingly, the strain degraded 1,4-dioxane faster after growth on THF than after growth on propane, but it degraded BCEE about three times faster after growth on propane than after growth on THF (32 versus 12 mg/h/g TSS). Although the products of BCEE and 1,3-dioxolane degradation were not analyzed, MTBE oxidation resulted in accumulation of tert-butyl alcohol that was not degraded further by the strain.
Microcosm studies.
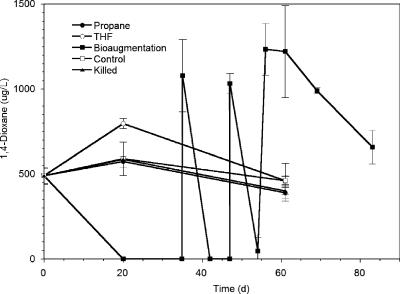
To test the utility of THF-degrading strains like strain ENV478 for treating 1,4-dioxane-contaminated aquifers, a microcosm study was conducted with field samples. THF-grown strain ENV478 cells were washed and added to triplicate microcosms (18 mg TSS/microcosm; final concentration, ∼5 × 108 cells/ml), and the microcosms were incubated aerobically for 80 days at 15°C (Fig. 5). 1,4-Dioxane (∼700 μg/liter) was rapidly degraded in the augmented microcosms before the first sampling time (day 20), and it was also rapidly degraded after repeated additions (∼1,000 μg/liter). After 53 days of incubation, fresh groundwater was added to the microcosms to replace the volume that had been removed for sampling. This process resulted in ∼30% dilution of the resident microbial population in the microcosms and in a concomitant decrease in 1,4-dioxane degradation. The 1,4-dioxane degradation activity in the microcosms recovered, however, and 1,4-dioxane continued to be degraded, albeit at a slightly lower rate. Little loss of 1,4-dioxane was observed in microcosms that did not receive strain ENV478 and in microcosms poisoned with HgCl2.
FIG. 5.
Biodegradation of 1,4-dioxane in aerobic aquifer microcosms. The different treatments are described in Materials and Methods. “Bioaugmentation” microcosms received THF-grown strain ENV478 (18 mg TSS each). The initial 1,4-dioxane concentration in the site samples was ∼500 μg/liter, and additional 1,4-dioxane was added to the bioaugmentation microcosms on days 35, 46, and 53. Fresh groundwater was added to the microcosms on day 53, which resulted in 30% dilution of the resident microbial population. The symbols indicate means (n = 3), and the error bars indicate standard errors.
DISCUSSION
Pseudonocardia sp. strains are widely distributed nocardioform actinomycetes that are abundant in many environments and are known to degrade a wide range of pollutants. Strain ENV478 and at least two other Pseudonocardia strains have now been shown to degrade the cyclic monoether THF (19, 28) and to cometabolically degrade 1,4-dioxane after growth on THF. P. dioxanivorans strain CB1190 (24, 28) was isolated from a THF enrichment and is the only Pseudonocardia strain that is known to grow on 1,4-dioxane as a sole carbon source. A related strain, Pseudonocardia sp. strain K1 (19), also grows on THF but not 1,4-dioxane and has been studied in detail, which has led to cloning and analysis of an apparent THF oxidation operon.
In this study, we isolated a new organism, strain ENV478, that is phylogenetically related to strains CB1190 and K1 (Fig. 1) but is not able to grow on 1,4-dioxane as a sole carbon source even though it appears that the same enzyme(s) in the strain is required to oxidize both substrates (Fig. 2). In this study we focused on evaluating why this strain cannot grow on 1,4-dioxane and on evaluating the use of this strain to degrade 1,4-dioxane in contaminated environments. The inability of strain ENV478 and other THF degraders to grow on 1,4-dioxane could be related to a number of factors, including the lack of induction of the requisite enzyme(s) by 1,4-dioxane or an inability to efficiently process metabolites for energy production.
THF degradation by Pseudonocardia strain K1 is facilitated by a plasmid-borne, NADH-dependent, multicomponent, binuclear iron THF monooxygenase (37). Consequently, growth on THF requires an expenditure of energy (for NADH generation) for the primary oxidation. Degradation of 1,4-dioxane by strain ENV478 was greatest after growth on THF, and the presence of THF inhibited degradation of 1,4-dioxane by this strain (Fig. 2). Furthermore, strain ENV478 appeared to accumulate 2HEAA, an expected product of a monooxygenation reaction with 1,4-dioxane, during 1,4-dioxane degradation. Therefore, we concluded that degradation of 1,4-dioxane and THF by strain ENV478 involves an initial oxidation of the cyclic ether by a monooxygenase that presumably is homologous to the THF monooxygenase of strain K1. Because strain ENV478 also can degrade propane, however, the presence of a short-chain alkane monooxygenase, some of which are known to oxidize ethers (31, 34), including possibly 1,4-dioxane (5), cannot be completely ruled out. However, the relatively low activity of propane-grown cells (Fig. 3) compared to the activity of THF-grown cells (Fig. 2) suggests that propane monooxygenase is a minor catalyst in this strain. Conversely, an alkane monooxygenase may be more important in BCEE oxidation by strain ENV478 because this organism degraded BCEE faster after growth on propane than after growth on THF.
The initial monooxidation of 1,4-dioxane likely results in production of 1,4-dioxane-2-ol, the hemiacetal of 2-hydroxyethoxy-2-acetaldehyde (Fig. 4). This compound could be oxidized by an alcohol dehydrogenase, as suggested previously for THF (37), or, because of the inherent instability of hemiacetals in water, could be chemically oxidized through the hydroxyaldehyde intermediate to 2HEAA. Because we observed that 2HEAA accumulated in cultures of Pseudomonas putida containing cloned toluene-4-monooxygenase genes and two propanotrophs, ENV421 and ENV425 (34; data not shown), it appears unlikely that production of 2HEAA requires multiple specific enzymatic reactions. More research is needed, however, to confirm this assumption. Although under some circumstances the 2HEAA might form PDX, this product is unlikely to occur in aqueous solutions, especially at the near-neutral pH of our cultures. Our attempts to isolate PDX from our cultures of ENV478 were unsuccessful, and even when we attempted to synthesize this compound in our laboratory, at pH >3 it was difficult to obtain PDX without 2HEAA present. The 1,4-dioxane biodegradation pathway in ENV478 (Fig. 4), therefore, appears to be similar to the 1,4-dioxane metabolic pathway proposed for rats (4).
Interestingly, in 1,4-dioxane biodegradation studies with a pure culture of fungi, Nakamiya et al. (27) observed only ethylene glycol, glycolic acid, and oxalic acid as 1,4-dioxane degradation products. This suggests either that an alternative degradation pathway was present in fungi, that 2HEAA was rapidly degraded to these products by the fungi and was not detected in the analyses performed in the study, or that the derivatization method used in the study did not allow detection of 2HEAA. Degradation of 2HEAA to these products could be facilitated by a dehydrogenase related to the diglycolic acid dehydrogenase of strain K1, which cleaves ether bonds adjacent to terminal carboxyl groups in both short-chain ethers (diglycolic acid) and long-chain polymers (polyethylene glycol) (41). A lack of a similar enzyme activity in ENV478 or a stringent substrate range that does not allow the enzyme to cleave 2HEAA, which may be the case in strain K1, may prevent these strains from metabolizing 2HEAA and result in their inability to grow on 1,4-dioxane. Parales et al. (28) suggested that long-term incubation of their THF-degrading cultures with THF and periodic additions of 1,4-dioxane may have led to a mutation that allowed strain CB1190 to grow on 1,4-dioxane. Our results suggest that such a mutation may have allowed the strain to metabolize 2HEAA, but to our knowledge, CB1190 cultures have never been analyzed for this 1,4-dioxane metabolite or for 2HEAA-metabolizing enzyme activity.
Despite the inability of strain ENV478 to grow on 1,4-dioxane in pure culture, this strain is still an effective biocatalyst, degrading 1,4-dioxane over an extended incubation period (∼80 days) (Fig. 5) in microcosms. The ability of this strain to continue to degrade 1,4-dioxane in microcosms may be due to its ability to utilize other carbon sources in the aquifer material to meet its energy and reductant demands for prolonged 1,4-dioxane degradation. Likewise, other microbes in the microcosms may transform the 2HEAA into products that are more readily used by strain ENV478. More research is needed to assess whether prolonged 1,4-dioxane degradation by strain ENV478 can be supported by exogenous carbon sources and to evaluate the fate of 2HEAA in environmental samples.
The inability of 1,4-dioxane to induce the THFmo homolog genes in strain ENV478 also could limit this strain's ability to grow on 1,4-dioxane by limiting the supply of enzyme needed to continuously process 1,4-dioxane. Expression of THFmo appears to be tightly regulated in strain K1 (37). Northern blot analysis demonstrated that the monooxygenase was produced during growth on THF but not after growth on succinate. Conversely, we demonstrated that strain ENV478 can degrade 1,4-dioxane even after growth on alternative substrates (Fig. 3), albeit at a rate lower than the rate observed for fully induced THF-grown cells. This suggests that the apparent THFmo of strain ENV478 is expressed constitutively and that the activity can be induced to even higher levels during growth on THF. The constitutively expressed enzyme may be enough to support prolonged degradation of 1,4-dioxane under certain conditions in the field, as we observed in our microcosms studies, in which 1,4-dioxane was degraded for >80 days (Fig. 5). The possibility that alternative unidentified inducers were present in the microcosm studies, however, cannot be ruled out (16, 31).
Poor induction of downstream enzymes also might limit the processing of 1,4-dioxane metabolites to support the growth of ENV478 on this compound. Although no genes that are obviously involved in 2HEAA metabolism are apparent in the cloned THF operon of strain K1, this strain does produce a diglycolic acid dehydrogenase that cleaves ether bonds adjacent to terminal carboxyl groups in short-chain ethers (41). The lack of such activity or the inability of 1,4-dioxane or its metabolites to induce this or similar activity in strain ENV478 also could limit 2HEAA metabolism. To test the ability of THF to induce 2HEAA metabolism by ENV478, cultures that had accumulated 2HEAA were fed THF. Although the added THF was readily degraded, no further degradation of 2HEAA was observed (data not shown), indicating that THF and its metabolites probably do not induce the expression of genes necessary for 2HEAA metabolism in strain ENV478. Additional studies are under way to further investigate the presence and induction of potential 2HEAA degradation genes in ENV478.
The ability of strain ENV478 to degrade 1,4-dioxane and other potentially important ether pollutants (BCEE, 1,3-dioxolane, and MTBE) warrants evaluation of its usefulness as a biocatalyst for in situ or ex situ treatment systems. In situ bioaugmentation with aerobic microorganisms has shown promise for treating recalcitrant pollutants (8, 11, 33, 35), but the technology has not been broadly accepted by the remediation community. Although the microcosms were inoculated with relatively large amounts of strain ENV478, the results of our microcosm study with strain ENV478 indicate that it may be possible to use this strains or similar strains to treat 1,4-dioxane contamination either in situ or in bioreactor systems. In studies of similar THF degraders in bioreactors the workers found that relatively large amounts of THF had to be added continuously to the systems to maintain 1,4-dioxane degradation activity (43). The ability of strain ENV478 to maintain 1,4-dioxane degradation activity in microcosms for >80 days without THF was, therefore, surprising and encouraging, but much additional work is needed to demonstrate the utility of such biocatalysts for remediating contaminated aquifers (33).
Acknowledgments
This work was supported by the Strategic Environmental Research and Development Program as part of project CU-1422.
REFERENCES
- 1.Adams, C. D., P. A. Scanlan, and N. D. Secrist. 1994. Oxidation and biodegradability enhancement of 1,4-dioxane using hydrogen peroxide and ozone. Environ. Sci. Technol. 28:1812-1818. [DOI] [PubMed] [Google Scholar]
- 2.Altschul, S. F., T. L. Madden, A. A. Scaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bernhardt, D., and H. Diekmann. 1991. Degradation of dioxane, tetrahydrofuran and other cyclic ethers by an environmental. Rhodococcus strain. Appl. Microbiol. Biotechnol. 36:120-123. [DOI] [PubMed] [Google Scholar]
- 4.Braun, W. H., and J. D. Young. 1977. Identification of γ-hydroxyethoxyacetic acid as the major urinary metabolite of 1,4-dioxane in the rat. Toxicol. Appl. Pharmacol. 39:33-38. [DOI] [PubMed] [Google Scholar]
- 5.Burback, B. L., and J. J. Perry. 1993. Biodegradation and biotransformation of groundwater pollutant mixtures by Mycobacterium vaccae. Appl. Environ. Microbiol. 59:1025-1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cole, J. R., B. Chai, R. J. Farris, Q. Wang, S. A. Kulam, D. M. Mcgarrell, G. M. Garrity, and M. Tiedje. 2005. The ribosomal database project (RDP-II): sequences and tools for high-throughput rRNA analysis. Nucleic Acids Res. 33:D294-D296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Daye, K. J., J. C. Groff, A. C. Kirpekar, and R. Mazumder. 2003. High efficiency degradation of tetrahydrofuran (THF) using a membrane bioreactor: identification of THF-degrading cultures of Pseudonocardia sp. strain M1 and Rhodococcus ruber isolate M2. J. Ind. Microbiol. Biotechnol. 30:705-714. [DOI] [PubMed] [Google Scholar]
- 8.DeFlaun, M. F., and R. J. Steffan. 2002. Bioaugmentation, p. 434-442. In G. Bitton (ed.), Encyclopedia of environmental microbiology. John Wiley & Sons, New York, N.Y.
- 9.Doddi, N., C. Versfelt, and D. Wasserman. October. 1977. Synthetic absorbable surgical devices of poly-dioxanone. U.S. patent 4,052,988.
- 10.Embley, T. M., J. Smida, and E. Stackebrandt. 1988. The phylogeny of mycolate-less wall chemotype IV actinomycetes and description of Pseudonocardiaceae fam. nov. Syst. Appl. Microbiol. 11:44-52. [Google Scholar]
- 11.Fathepure, B. Z., V. K. Elango, H. Singh, and M. A. Bruner. 2005. Bioaugmentation potential of a vinyl chloride-assimilating Mycobacterium sp., isolated from a chloroethene-contaminated aquifer. FEMS Microbiol. Lett. 248:227-234. [DOI] [PubMed] [Google Scholar]
- 12.Fincher, E. L., and W. J. Payne. 1962. Bacterial utilization of ether glycols. Appl. Microbiol. 10:542-547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Harland, W., R. L. Crawford, P. J. Chapman, and S. Dagley. 1975. Metabolic function and properties of 4-hydroxyphenylacetic acid 1-hydroxylase from Pseudomonas acidovorans. J. Bacteriol. 121:272-285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jackson, R. E., and V. Dwarakanath. 1999. Chlorinated degreasing solvents: physical-chemical properties affecting aquifer contamination and remediation. Ground Water Monit. Remediation 19:102-110. [Google Scholar]
- 15.Johnson, E. L., C. A. Smith, K. T. O'Reilly, and M. R. Hyman. 2004. Induction of methyl tertiary butyl ether (MTBE)-oxidizing activity in Mycobacterium vaccae JOB5 by MTBE. Appl. Environ. Microbiol. 70:1023-1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Johnson, J. L. 1994. Similarity analysis of rRNAs, p. 683-700. In P. Gerhardt, R. G. E. Murray, W. A. Wood, and N. R. Krieg (ed.), Methods for general and molecular bacteriology. American Society for Microbiology, Washington, D.C.
- 17.Kampfer, P., and R. M. Kroppenstedt. 2004. Pseudonocardia benzenivorans sp. nov. Int. J. Syst. Evol. Microbiol. 54:749-751. [DOI] [PubMed] [Google Scholar]
- 18.Klečka, G. M., and S. J. Gonsior. 1986. Removal of 1,4-dioxane from wastewater. J. Hazard. Mater. 13:161-168. [Google Scholar]
- 19.Kohlweyer, U., B. Thiemer, T. Schrader, and J. R. Andreesen. 2000. Tetrahydrofuran degradation by a newly isolated culture of Pseudonocardia sp. strain K1. FEMS Microbiol. Lett. 186:301-306. [DOI] [PubMed] [Google Scholar]
- 20.Kumar, S., K. Tamura, I. B. Jakobsen, and M. Nei. 2001. MEGA2: molecular evolutionary genetics analysis software. Bioinformatics 17:1244-1245. [DOI] [PubMed] [Google Scholar]
- 21.Lee, S. D., E. S. Kim, and Y. C. Hah. 2000. Phylogenetic analysis of the genera Pseudonocardia and Actinobispora based on 16S ribosomal DNA sequences. FEMS Microbiol. Lett. 182:125-129. [DOI] [PubMed] [Google Scholar]
- 22.Lee, S. D., E. S. Kim, K. L. Min, W. Y. Lee, S. O. Kang, and Y. C. Hah. 2001. Pseudonocardia kongjuensis sp. nov., isolated from a gold mine cave. Int. J. Syst. Evol. Microbiol. 51:1505-1510. [DOI] [PubMed] [Google Scholar]
- 23.Lesage, S., R. E. Jackson, M. W. Priddle, and P. G. Reimann. 1990. Occurrence and fate of organic solvent residues in anoxic groundwater at the Gloucester Landfill, Canada. Environ. Sci. Technol. 24:559-566. [Google Scholar]
- 24.Mahendra, S., and L. Alvarez-Cohen. 2005. Pseudonocardia dioxanivorans sp. nov., a novel actinomycete that grows on 1,4-dioxane. Int. Syst. Evol. Microbiol. 55:593-598. [DOI] [PubMed] [Google Scholar]
- 25.McVeigh, H. P., J. Munro, and T. M. Embley. 1994. The phylogenetic position of Pseudoamycolata halophobica (Akimov et al. 1989) and a proposal to reclassify it as Pseudonocardia halophobica. Int. J. Syst. Bacteriol. 44:300-302. [DOI] [PubMed] [Google Scholar]
- 26.Mohr, T. K. G. 2001. Solvent stabilizers white paper. [Online.] Santa Clara Valley Water District, San Jose. Calif. http://www.valleywater.org/Water/Water_Quality/Protecting_your_water/_Solvents/_PDFs/SolventStabilizers.pdf.
- 27.Nakamiya, K., S. Hashimoto, H. Ito, J. Edmonds, and M. Morita. 2005. Degradation of 1,4-dioxane and cyclic ethers by an isolated fungus. Appl. Environ. Microbiol. 71:1254-1258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Parales, R. E., J. E. Adanus, N. White, and H. D. May. 1994. Degradation of 1,4-dioxane by an actinomycete in pure cultures. Appl. Environ. Microbiol. 60:4527-4530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Priddle, M. W., and R. E. Jackson. 1991. Laboratory column measurement of VOC retardation factors and comparison with field values. Ground Water 29:260-266. [Google Scholar]
- 30.Reichert, K., A. Lipski, S. Pradella, E. Stackebrandt, and K. Altendorf. 1998. Pseudonocardia asaccharolytica sp. nov. and Pseudonocardia sulfidoxydans sp. nov., two new dimethyl disulfide-degrading actinomycetes and emended description of the genus Pseudonocardia. Int. J. Syst. Bacteriol. 48:441-449. [DOI] [PubMed] [Google Scholar]
- 31.Smith, C. A., K. T. O'Reilly, and M. R. Hyman. 2003. Characterization of the initial reactions during the cometabolic degradation of methyl tert-butyl ether (MTBE) by propane-grown Mycobacterium vaccae JOB5. Appl. Environ. Microbiol. 69:796-804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stefan, M. I., and J. R. Bolton. 1998. Mechanism of the degradation of 1,4-dioxane in dilute aqueous solutions using the UV/hydrogen peroxide process. Environ. Sci. Technol. 32:1588-1595. [Google Scholar]
- 33.Steffan, R. J., K. L. Sperry, M. T. Walsh, S. Vainberg, and C. W. Condee. 1999. Field-scale evaluation of in situ bioaugmentation for remediation of chlorinated solvents in groundwater. Environ. Sci. Technol. 33:2771-2781. [Google Scholar]
- 34.Steffan, R. J., K. McClay, S. Vainberg, C. W. Condee, and D. Zhang. 1997. Biodegradation of the gasoline oxygenates methyl tert-butyl ether, ethyl tert-butyl ether, and tert-amyl methyl ether by propane-oxidizing bacteria. Appl. Environ. Microbiol. 63:4216-4222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Steffan, R. J., Y. Farhan, C. W. Condee, and S. Drew. 2003. Bioremediation at a New Jersey site using propane-oxidizing bacteria, p. 503-516. In E. Moyer and P. Kostecki (ed.), MTBE remediation handbook. Amherst Scientific Publisher, Amherst Mass.
- 36.Thiemer, B., J. R. Andreesen, and T. Schrader. 2001. The NADH-dependent reductase of a putative multicomponent tetrahydrofuran mono-oxygenase contains a covalently bound FAD. Eur. J. Biochem. 268:3774-3782. [DOI] [PubMed] [Google Scholar]
- 37.Thiemer, B., J. R. Andreesen, and T. Schrader. 2003. Cloning and characterization of a gene cluster involved in tetrahydrofuran degradation in Pseudonocardia sp. strain K1. Arch. Microbiol. 179:266-277. [DOI] [PubMed] [Google Scholar]
- 38.U.S. Environmental Protection Agency. 1998. U.S. EPA test methods for evaluating solid waste, physical/chemical methods SW846, 3rd edition, revision 5. U.S. Environmental Protection Agency, Washington, D.C.
- 39.U.S. Environmental Protection Agency. 1999. U.S. EPA methods and guidance for analysis of water and wastes. Publication EPA-600/4-79-020. U.S. Environmental Protection Agency, Washington, D.C.
- 40.Woo, Y. T., J. C. Arcos, M. F. Argus, G. W. Griffin, and K. Nishiyama. 1977. Structural identification of p-dioxane-2-one as the major urinary metabolite of p-dioxane. Naunyn-Schmiedeberg's Arch. Pharmacol. 299:283-287. [DOI] [PubMed] [Google Scholar]
- 41.Yamashita, M., A. Tani, and F. Kawai. 2005. A new ether bond-splitting enzyme found in gram-positive polyethylene glycol 6000-utilizing bacterium, Pseudonocardia sp. Appl. Microbiol. Biotechnol. 66:174-179. [DOI] [PubMed] [Google Scholar]
- 42.Young, J. D., W. H. Braun, and P. J. Gehring. 1978. The dose-dependent fate of 1,4-dioxane in rats. J. Environ. Pathol. Toxicol. 2:263-282. [PubMed] [Google Scholar]
- 43.Zenker, M. J., R. C. Borden, and M. A. Barlaz. 2000. Mineralization of 1,4-dioxane in the presence of a structural analog. Biodegradation 11:239-246. [DOI] [PubMed] [Google Scholar]