Abstract
In the ubiquitous marine bacterium Pseudoalteromonas tunicata, subpopulations of cells are killed by the production of an autocidal protein, AlpP, during biofilm development. Our data demonstrate an involvement of this process in two parameters, dispersal and phenotypic diversification, which are of importance for the ecology of this organism and for its survival within the environment. Cell death in P. tunicata wild-type biofilms led to a major reproducible dispersal event after 192 h of biofilm development. The dispersal was not observed with a ΔAlpP mutant strain. Using flow cytometry and the fluorescent dye DiBAC4(3), we also show that P. tunicata wild-type cells that disperse from biofilms have enhanced metabolic activity compared to those cells that disperse from ΔAlpP mutant biofilms, possibly due to nutrients released from dead cells. Furthermore, we report that there was considerable phenotypic variation among cells dispersing from wild-type biofilms but not from the ΔAlpP mutant. Wild-type cells that dispersed from biofilms showed significantly increased variations in growth, motility, and biofilm formation, which may be important for successful colonization of new surfaces. These findings suggest for the first time that the autocidal events mediated by an antibacterial protein can confer ecological advantages to the species by generating a metabolically active and phenotypically diverse subpopulation of dispersal cells.
Most bacteria in the environment live in biofilms rather than as free-living cells (8). Bacteria in biofilms live in tightly associated, matrix-encased groups attached to a surface (8, 18). Biofilms can contaminate a broad range of environmental, industrial, and biomedical surfaces (30) and are recognized as the cause of many chronic human infections (8, 9). They can also act as reservoirs for pathogenic bacteria, such as Vibrio cholerae and Legionella pneumophila, in drinking water systems and within the environment (38, 50, 59). Biofilms exhibit a marked increase in resistance to environmental stress (for example, protozoan grazing, antibiotics, or host immune responses) compared to free-living single bacteria and often produce a protective extracellular matrix (6, 58) that also facilitates cell-cell interactions, including the exchange of chemical signal molecules.
In particular, two behaviors of bacteria in biofilms are now a focus of attention because of their ability to greatly enhance the survival and spread of bacteria within the environment. These processes are phenotypic variation and dispersal. Cells dispersing from biofilms often exhibit a high degree of phenotypic and genotypic variation that enhances survival in the face of changing environmental conditions (4, 21, 31). In addition, cells within biofilms can at times disperse in order to colonize new surfaces (5, 17, 28, 53). Active processes are often used by bacteria to promote dispersal; these include, for example, enzyme-mediated breakdown of the biofilm matrix (5, 28, 29, 33) or the production of surfactants which loosen cells from the biofilm (10). Dispersal processes may be under the control of cell-cell communication systems that facilitate coordinated group behavior of bacteria in biofilms (49), and changes in nutrient availability have also been linked to biofilm dispersal (19, 25, 53). Biofilm development is thus a dynamic process in which phenotypic variation and dispersal are key factors that influence biofilm function within the environment.
Pseudoalteromonas tunicata is an epiphytic marine bacterium that has been isolated and detected on higher organisms in different marine habitats (15, 23, 24, 55). P. tunicata produces a suite of antimicrobial compounds thought to provide a competitive advantage to the organism during biofilm formation on crowded surfaces (34). Killing in P. tunicata biofilms occurs through the activity of AlpP, a large (190-kDa) autolytic protein (34). Several P. tunicata mutants have been generated, including ΔAlpP (34), D2W2, and D2W3 (13, 14), which are deficient in the production of AlpP and show no cell death during biofilm development. The lack of cell death occurs despite the fact that the same biofilm architecture is maintained, as determined by statistical analyses of three-dimensional confocal microscopy images (22, 34). Because mutants that do not undergo cell death can be generated, we hypothesized that these events may represent an evolved capacity of significance to biofilm development.
Here, we report that cell death during biofilm development of P. tunicata is involved in key processes of importance to the survival and spread of bacterial biofilms within the environment. The findings reported in this paper on variation in colonization traits of dispersal cells suggest that the self-lysis in microcolony biofilms enhances the ability of other cells to disperse from within biofilms and colonize new surfaces.
MATERIALS AND METHODS
Bacterial strains and media.
P. tunicata was routinely cultivated at room temperature in Väätänen nine-salt solution (VNSS) (35) supplemented with streptomycin (100 μg ml−1) and kanamycin (50 μg ml−1) for the P. tunicata ΔAlpP mutant (34). Biofilms were grown in marine minimal medium (45) containing 0.01% trehalose.
Biofilm cultures.
P. tunicata wild-type and ΔAlpP mutant strains were grown in continuous-culture flow cells (channel dimensions, 1 by 4 by 40 mm) at room temperature as previously described (42). Channels were inoculated with 0.5 ml of early-stationary-phase cultures containing approximately 1 × 109 cells ml−1 and incubated without flow for 1 h at room temperature. Flow was then started with a mean flow velocity of 0.2 mm s−1 in the flow cells, corresponding to laminar flow with a Reynolds number of 0.02.
To characterize biofilm dispersal, the number of viable bacteria in the effluent of three independent biofilm channels of both P. tunicata wild-type and ΔAlpP mutant biofilms was determined by serial plate counts using VNSS.
To determine whether AlpP-mediated killing induces dispersal and increases phenotypic variation among dispersal cells (see below), we added purified AlpP to three biofilm channels of 144-h ΔAlpP mutant biofilms. AlpP was prepared and purified from the supernatant as previously described (27). AlpP (10 to 12 μg in dialysis buffer; 20 mM Tris, 0.3 M NaCl) was injected into the flow cells by use of a syringe needle. Silicone tubing at either side of the flow cell was then blocked off with tubing clamps. As a control, dialysis buffer (with or without the addition of heat-inactivated AlpP) was inoculated into separate flow-cell channels. Biofilms were incubated at 25°C for 5 h before the flow was turned on, and effluent was collected at 30-min intervals. Effluent from both control and AlpP add-back channels was spread onto VNSS plates in serial dilutions to quantify the biofilm dispersal of viable cells. Phenotypic variations in motility, growth, and biofilm formation were measured as described below.
Measurement of phenotypic variation.
To investigate the hypothesis that cell lysis within microcolonies promotes phenotypic variation in biofilms, effluent was spread plated onto VNSS, LB10, Marine agar (Difco, Becton Dickinson), and Davis minimal medium (Difco, Becton Dickinson) in order to detect colony morphology variation. In addition, 20 colonies derived from wild-type and ΔAlpP mutant biofilms were randomly selected from VNSS agar and screened for growth, biofilm formation, and motility. Three time points were chosen: 24 h (before the onset of cell death), 72 h (shortly after the onset of cell death), and 144 h (when cell death had extended throughout the biofilm). Overnight cultures (15 μl) of the 20 colonies were inoculated into 1.5 ml of fresh VNSS in 24-well tissue culture plates. Plates were incubated at 25°C with agitation (130 rpm). The optical density at 600 nm was measured after 24 h as an indicator for growth ability. To measure biofilm-forming ability, wells of the tissue culture plates were stained with crystal violet for 20 min. After the wells were washed twice with NSS, crystal violet was extracted in 95% ethanol, and the absorbance was read at 600 nm. As an indicator for swimming motility, the variants were stab inoculated into 0.4% VNSS agar, and the growth radius was measured after 24 h.
To provide evidence that the measured variation is a biofilm-specific trait, the wild-type and ΔAlpP mutant strains were inoculated into 500-ml planktonic cultures in marine minimal medium with agitation (130 rpm). Aliquots were taken at 24 h, 72 h, and 144 h and spread onto VNSS. Twenty colonies were randomly selected and subjected to the same characterization process as described above.
To determine the relative variation among the wild-type and ΔAlpP mutant strains for each time point, a statistical coefficient of variation (CV) was calculated. The CV is the standard deviation divided by the mean and multiplied by 100. This gives a percentage value indicating the variability around the mean in relation to the size of the mean.
A variance analysis (F test) was performed to determine whether a difference in variation was statistically significant. The F test can be used to determine whether variances of two samples are different. For a sample size of 20 and a 95% confidence level, the F value should be >2.1 to reject the hypothesis that the two variances are equal.
Fluorescent staining.
The membrane potential probe bis-(1,3-dibutylbarbituric acid)trimethine oxonol [DiBAC4(3); Molecular Probes, Inc.] was used to investigate whether cell death influences the metabolic activity of dispersal cells. DiBAC4(3) is a green fluorescent anionic dye commonly used in flow cytometry studies as an indicator of membrane potential. It enters the cell as a result of membrane depolarization (occurring in dead cells or metabolically inactive cells) and binds to intracellular proteins or membranes which thereby exhibit green fluorescence. Increased depolarization results in a greater influx of the anionic dye and thus results in an increased green fluorescence. Active cells with a charged membrane largely exclude the dye and remain less fluorescent (7). Effluent of both wild-type and ΔAlpP mutant biofilms at 72 h (shortly after the onset of cell death) and 144 h (with extensive cell death throughout the wild-type biofilm) was stained according to the manufacturer's description. As controls, P. tunicata cells in logarithmic growth phase were stained. One half of the control cells were not treated (active cells), and the other half were heat killed for 10 min at 70°C (nonactive cells) before staining.
Flow cytometry.
Flow cytometric analysis was performed using a BD FACSCalibur sort flow cytometer (BD Biosciences, Sydney, Australia) equipped with an air-cooled 100 mW argon ion laser (488 nm) for excitation. The positive and negative controls were analyzed first, and the threshold was set to just below the population of bacteria on a bivariate dot plot of side scatter (SSC) versus forward scatter (FSC). A gate was defined around this control population, and the fluorescence was monitored on a histogram of green florescence (FL1). All samples were run in triplicate with 10,000 cells being analyzed on a histogram of green fluorescence. Data analysis was carried out using Summit offline software (Cytomation, Inc). Positive and negative controls were analyzed first to ensure that both populations were separated on the FL1 histogram (green fluorescence detector). For all samples, a region around the main bacterial population in the bivariate dot plot of SSC versus FSC was defined, and only cells within this region were analyzed in the univariate FL1 histogram. Regions around active (R1) and depolarized (R2) cells on the univariate FL1 histogram were then defined. The percentage of cells in each region was recorded, and the mean value and standard deviation were calculated from triplicate samples. An unpaired, two-sided t test was performed to test the hypothesis that the analyzed samples differed significantly.
RESULTS
AlpP-mediated cell death is required for biofilm dispersal events.
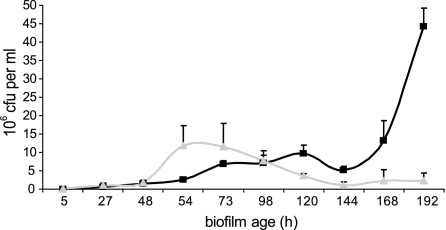
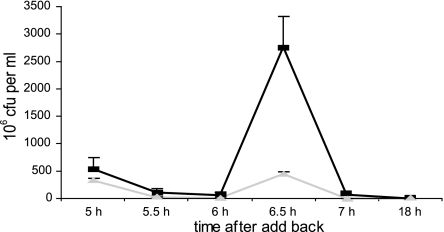
In the initial report on cell death in P. tunicata biofilms, it was found that death and lysis of a subpopulation of cells occur in the center of microcolonies after 48 h of biofilm formation (34). In this study, we investigated whether AlpP-mediated cell death plays a role in P. tunicata biofilm dispersal. Numbers of viable dispersal cells in the biofilm effluent from P. tunicata wild-type and ΔAlpP mutant strains were quantified and statistically compared over a period of 192 h (8 days) of biofilm development. Both strains showed little dispersal for the first 48 h of biofilm development with 1.51 × 106 wild-type CFU ml−1 and 1.86 × 106 ΔAlpP mutant CFU ml−1 in the biofilm effluent (Fig. 1). The P. tunicata wild-type strain showed a steady increase in viable cells dispersing from the biofilm over the time course of the experiment; there was a major detachment event at 192 h, when most of the biofilm was removed from the substratum. This detachment event correlates with extensive AlpP-mediated killing within the biofilm. At this time point, the effluent contained as much as 4.43 × 107 CFU ml−1. In contrast, the ΔAlpP mutant did not show a major biofilm detachment event, and dispersal remained low over the time course of the experiment, with only 2.28 × 106 CFU ml−1 dispersing at 192 h. Several replicate experiments over longer periods of time (up to 240 h) failed to show a large dispersal event for the ΔAlpP mutant strain. Effluent CFU counts always remained below 4 × 106 CFU ml−1. However, when purified AlpP was added back to mature ΔAlpP mutant biofilms, a major dispersal event was induced (Fig. 2). After AlpP add-back, the ΔAlpP mutant showed a 10-fold increase in the number of dispersal cells compared to the Tris buffer control (Fig. 2). This effect was not observed when heat-inactivated AlpP protein (in Tris buffer) was added to the mutant biofilms (data not shown). The reduction in CFU subsequent to a major dispersal event is due to the complete loss of the biofilm, which is similar to what occurs after the main dispersal event of the wild-type biofilm past 192 h (Fig. 1). The nature of the add-back experiment, i.e., stopping the flow for 5 h, led to a considerably higher number of CFU in the effluent of both control and add-back channels than that in the experiment shown in Fig. 1. Three independent experiments revealed similar results.
FIG. 1.
Biofilm dispersal shown in effluent viable counts (CFU). P. tunicata wild-type (black squares) shows a significant sloughing event at 192 h. No major increase in dispersal can be detected for the ΔAlpP mutant (grey triangles). Error bars represent the standard deviations for three independent experiments.
FIG. 2.
ΔAlpP mutant biofilm dispersal after AlpP add-back (black squares) and buffer control (20 mmol Tris) (grey triangles). Dispersal is shown with effluent viable counts 6 to 8 h after inoculation of AlpP and Tris, respectively. Error bars represent the standard deviations for three independent experiments.
Cell death during biofilm development enhances metabolic activity of surviving dispersal cells.
We studied cell activity of the dispersal population of both strains at two different time points during biofilm formation (72 h and 144 h) using the fluorescent dye DiBAC4(3) in conjunction with flow cytometry. DiBAC4(3) exhibits enhanced fluorescence when it enters cells with depolarized membranes. At 72 h, dispersal cells of both the wild type and the ΔAlpP mutant showed two distinct cell populations (Fig. 3A and B). The subpopulation with lower fluorescence corresponded to active cells with polarized membranes, and the subpopulation with higher fluorescence corresponded to cells with reduced activity (depolarized membranes). At 72 h, in both strains, a large proportion of dispersal cells were active and a smaller fraction of the population had depolarized membranes (Fig. 3A and B). At 144 h, the ΔAlpP mutant dispersal population consisted mainly of cells with depolarized membranes, and only a very small population of active cells could be detected (Fig. 3D). In contrast, at 144 h of biofilm development, when large regions of lysis occurred, wild-type dispersal cells still demonstrated two distinct fluorescent peaks, showing a large active cell population as well as a population of cells with depolarized membranes (Fig. 3C).
FIG. 3.
Flow cytometry analysis of biofilm effluent. Cells were stained with the green membrane potential probe DiBAC4(3) at 72 h (wild type [A] and ΔAlpP mutant [B]) and at 144 h (wild type [C] and ΔAlpP mutant [D]) of biofilm development. The histograms show green fluorescence (FL1-H in relative fluorescence units), with R1 corresponding to active cells and R2 corresponding to cells with depolarized membranes. Mean counts and standard deviations for each region as a percentage of total event counts were calculated from triplicate samples.
A t test was performed to establish whether the variability between subpopulation phenotypes observed at 72 h and 144 h was significant. The results demonstrated that the difference between the wild-type and ΔAlpP mutant active subpopulations was highly significant (P = 4.9 × 10−5). Similarly, the differences between the depolarized subpopulations at the two time points also revealed a highly significant P value, 5.2 × 10−5. It confirmed that the subpopulations observed at 144 h were significantly different from those observed at the 72-h time point.
Cell death is linked to phenotypic variation of biofilm dispersal cells.
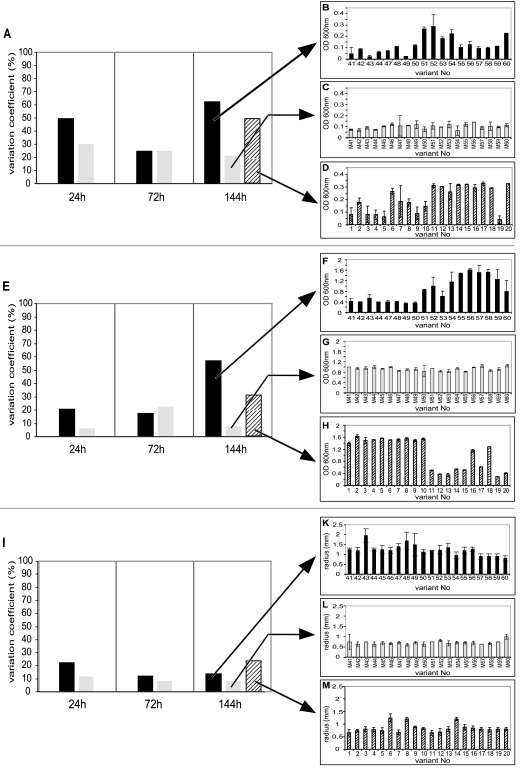
Many organisms show enhanced phenotypic variation during biofilm growth (1, 12, 43, 46, 51, 61). We investigated the occurrence of morphological colony variants during biofilm formation by plating biofilm effluent onto several different medium types over 10 days of biofilm formation. Unexpectedly, we found that no biofilm-specific morphological colony variants could be detected for the wild type or the ΔAlpP mutant, as all colonies had normal wild-type appearance at all investigated time points. However, we also investigated whether variation among dispersal cells may occur in phenotypic traits other than colony morphology—in particular, traits that are relevant for colonization of new surfaces. We therefore screened from VNSS agar plates 20 randomly picked colonies derived from biofilms of both strains for their growth ability, biofilm formation, and motility. We observed that colonies from the wild type showed very high variation in their biofilm-forming ability and growth ability (Fig. 4A, B, E, and F). The highest variation was detected at the latest time point of biofilm development (144 h), when biofilm killing was well established. At this time point, wild-type phenotypic variation ranked as high as 62% for biofilm formation and 57% for growth. Colonies derived from ΔAlpP mutant biofilms did not differ greatly in all three investigated traits, and variation at 144 h was only 21.02% for biofilm formation and 7.56% for growth (Fig. 4A, C, E, and G). Variations in motility of biofilm dispersal cells were comparably low for the two strains (Fig. 4I). However, a small number (0.03%) of wild-type colonies showed an interesting phenotype with significantly decreased biofilm formation as well as increased motility. In repeat experiments, this phenotype was not detected in the ΔAlpP mutant colonies (data not shown).
FIG. 4.
Variation in biofilm formation (A to D), growth (E to H), and motility (I to M) of P. tunicata wild-type (black bars) and ΔAlpP mutant (grey bars) biofilm dispersal cells. Variation coefficients (%) are calculated for 20 colonies after 24 h, 72 h, and 144 h of biofilm growth. The white bars with black stripes show variation for ΔAlpP mutant biofilm dispersal cells after AlpP add-back at 144 h. Error bars represent the standard deviations for three independent experiments.
An F test analysis showed that differences in variance between the wild type and ΔAlpP mutant were significant, i.e., with an F value of >2.1, for biofilm formation at 144 h (F = 14.2), for growth at 24 h (F = 11.4) and at 144 h (F = 46.0), and for motility at 24 h (F = 12.4), 72 h (F = 4.9), and 144 h (F = 9.3).
To investigate whether the high variation of wild-type biofilm dispersal cells is caused by AlpP-mediated cell death, purified AlpP protein was added back to mature ΔAlpP mutant biofilms, and dispersal cells were tested for variation. Interestingly, we observed that phenotypic diversity increased in a manner similar to that of the wild-type strain. Variation in growth rate increased from 7.6% to 31%, variation in biofilm formation from 21% to 49.6%, and variation in motility from 8.2% to 23.9% after AlpP add-back (Fig. 4A, D, E, H, I, and M).
Furthermore, the question of whether the high variation of wild-type dispersal cells occurs specifically in biofilms (and not planktonic cells) was examined. Thus, wild-type and ΔAlpP mutant colonies deriving from nonbiofilm batch cultures were tested for variations in the same traits. However, levels of variation in wild-type planktonic cells were found to be significantly lower than those of wild-type biofilm cells and, likewise, levels of variation in the ΔAlpP mutant planktonic cells were lower than those of ΔAlpP mutant biofilm cells for all three traits (data not shown).
DISCUSSION
Because of their group structure and function, biofilms are often considered primitive multicellular systems (47, 60). In multicellular organisms, the self-destruction of surplus or damaged cells is a normal component of development. More primitive forms of self-destruction are also observed among specialized differentiating bacteria, such as Myxococcus xanthus or Streptomyces spp. (16, 64). Recently, striking patterns of self-lysis have been observed during biofilm formation of several bacteria that are classically viewed as “nondifferentiating” organisms, including Pseudomonas aeruginosa (62), Vibrio cholerae (D. McDougald, J. S. Webb, and S. Kjelleberg, unpublished data), and P. tunicata (34). The ecological role of cell death in these nondifferentiating bacteria is not yet understood. Neither is it clear whether cell death occurs as part of a regulated “program” of biofilm development or simply results from the response of individual cells to changes in their immediate environment. However, one hypothesis underlying our research is that many fundamental behaviors observed in biofilms represent evolutionary precursors of processes observed in higher biological systems.
Several studies have observed patterns of gene expression that are specifically associated with different developmental stages of biofilm development (11, 52, 56). However, it has also been shown that hydrodynamics, nutrient load, and intracellular carbon flux have major impacts, presumably by altering the expression of cellular traits essential for bacterial adaptation during the different stages of biofilm formation (9, 32, 57, 63). Thus, current models for biofilm formation include both that of a developmental life cycle (56) and that of biofilm behaviors that result simply from the cumulative effect of single cells responding to changes within their environment (32).
It has not yet been established whether AlpP-mediated cell death is a programmed event during biofilm development. However, our study shows that these self-lysis events can confer certain ecological advantages to a subpopulation of surviving cells. In particular, the results of the present study reveal that self-induced cell lysis plays an important role in the dispersal process of P. tunicata biofilms. A metabolically active dispersal population with high phenotypic variation was found to be generated from P. tunicata wild-type biofilms after cell death had occurred but was not observed for the AlpP mutant defective in autolysis (Fig. 3 and 4).
Mechanisms by which biofilms regulate dispersal are becoming an increasing focus of research and include quorum sensing signals (49), exopolysaccharide-degrading enzymes (5, 10), nutrient levels (25, 53), the cell division cycle (2), and the carbon storage regulator (CsrA) (26). Interestingly, biofilm cells are often observed to disperse from within microcolonies, and the formation of cavities within microcolonies (hollow microcolonies) often precedes the dispersal process in several organisms (3, 10, 29, 52, 62). Similarly, for P. tunicata, it may be hypothesized that regions of lysis cause a weakening of the biofilm structure through the formation of hollow microcolonies and the disruption of cell aggregates, leading to dispersal. Furthermore, viable cells within the region of lysis are often observed to appear larger and more brightly fluorescent (data not shown), suggesting that lysis material from dead cells can also contribute to the nutrient support of surviving cells, which then disperse with an increased metabolic activity. The use of dead cells as a nutrient source for surviving bacteria has also been proposed to occur during differentiation processes in other organisms, including sporulation of Bacillus subtilis (20), mycelium formation of Streptomyces sp. (40, 41), and biofilm formation of Staphylococcus aureus (48). Here, we hypothesize that cell death and lysis in P. tunicata biofilms causes a weakening of the biofilm structure and adds to the nutrient supply of surviving cells, leading to a metabolically active dispersal population.
It is generally recognized that a high diversity within a community protects against unfavorable conditions by increasing the range of conditions in which a community as a whole can thrive (39, 54). As seen in this study, in P. tunicata biofilms, the diversity affected three different traits important for survival and colonization of new surfaces: motility, growth ability, and biofilm formation. Interestingly, levels of variation for the three traits investigated appear to be relatively stable in the dispersal cells, as three culturing steps in culture medium did not allow reversion of the phenotypes. The highest variation was detected in the wild type after cell death had occurred, with some variants showing high growth rates and rapid biofilm formation and some showing slow growth rates and being mostly deficient in biofilm formation (Fig. 4B and F). Furthermore, some variants derived from wild-type biofilms showed increased motility (Fig. 4K). Each of these phenotypes may influence the ability of P. tunicata cells to colonize surfaces under different environmental conditions. For example, a high growth and biofilm formation rate may enhance colonization under high nutrient availability and, conversely, a slow growth rate and decreased biofilm formation may promote colonization under nutrient-limited conditions. A phenotype with increased motility could enhance settlement at more distant surfaces, which could lead to a wider distribution of the organism. The need to disperse to new habitats is an important constraint on all organisms with sessile stages in their life cycle. Interestingly, the generation of variability among dispersal propagules has been observed for many sessile colonizing eukaryotes (36, 37, 44). It is well established for these organisms that variation in a range of phenotypes within propagule populations is a strategy used to ensure successful colonization of new surfaces in different habitats. For example, differences in swimming ability due to variations in the size and nutritional status of larvae of the bryozoans Bugula neritina and Watersipora subtorquata and the ascidian Diplosoma listerianum lead to variations in settlement distances (37).
Recently, it was proposed that the generation of phenotypically different and stable dispersal cells in the model biofilm-forming bacterium P. aeruginosa follows phage-mediated death of a subpopulation of cells (61, 62). Although the mechanisms for inducing variation in P. tunicata biofilms are yet to be fully investigated, the antibacterial and autolytic protein, AlpP, is clearly involved, seeing as add-back of purified AlpP induced cell death and dispersal in mature ΔAlpP mutant biofilms as well as increased phenotypic variation among dispersal cells.
Our findings suggest that developmental cell death and its consequences confer ecological advantages to groups of bacteria in P. tunicata biofilms and, more generally, the fitness of the species by generating a diverse but stable dispersal population. It remains to be established whether these benefits occur indirectly or as part of a regulated genetic program in P. tunicata. However, because patterns of cell death during biofilm development are a feature of many bacteria, exploring the role of self-induced lysis and generation of phenotypically different dispersal cells can lead to a better understanding of the ecology of the sessile lifestyle of nondifferentiating bacteria as well as the development of potential control mechanisms of bacterial biofilms.
Acknowledgments
We thank our colleagues at the University of New South Wales for their support and René Prochnow for help with the illustrations.
This research was supported by grants from the Australian Research Council and the Centre for Marine Biofouling and Bio-Innovation at the University of New South Wales, Australia. B.C.F. was funded by a Macquarie University Research Fellowship, Macquarie University, Australia.
REFERENCES
- 1.Ali, A., M. H. Rashid, and D. K. Karaolis. 2002. High-frequency rugose exopolysaccharide production by Vibrio cholerae. Appl. Environ. Microbiol. 68:5773-5778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Allison, D. G., D. J. Evans, M. R. Brown, and P. Gilbert. 1990. Possible involvement of the division cycle in dispersal of Escherichia coli from biofilms. J. Bacteriol. 172:1667-1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boles, B. R., M. Thoendel, and P. K. Singh. 2005. Rhamnolipids mediate detachment of Pseudomonas aeruginosa from biofilms. Mol. Microbiol. 57:1210-1223. [DOI] [PubMed] [Google Scholar]
- 4.Boles, B. R., M. Thoendel, and P. K. Singh. 2004. Self-generated diversity produces “insurance effects” in biofilm communities. Proc. Natl. Acad. Sci. USA 101:16630-16635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boyd, A., and A. M. Chakrabarty. 1994. Role of alginate lyase in cell detachment of Pseudomonas aeruginosa. Appl. Environ. Microbiol. 60:2355-2359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Branda, S. S., S. Vik, L. Friedman, and R. Kolter. 2005. Biofilms: the matrix revisited. Trends Microbiol. 13:20-26. [DOI] [PubMed] [Google Scholar]
- 7.Brauner, T., D. F. Hulser, and R. J. Strasser. 1984. Comparative measurements of membrane potentials with microelectrodes and voltage-sensitive dyes. Biochim. Biophys. Acta 771:208-216. [DOI] [PubMed] [Google Scholar]
- 8.Costerton, J. W., Z. Lewandowski, D. DeBeer, D. Caldwell, D. Korber, and G. James. 1994. Biofilms, the customized microniche. J. Bacteriol. 176:2137-2142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Costerton, J. W., P. S. Stewart, and E. P. Greenberg. 1999. Bacterial biofilms: a common cause of persistent infections. Science 284:1318-1322. [DOI] [PubMed] [Google Scholar]
- 10.Davey, M. E., N. C. Caiazza, and G. A. O'Toole. 2003. Rhamnolipid surfactant production affects biofilm architecture in Pseudomonas aeruginosa PAO1. J. Bacteriol. 185:1027-1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.De Kievit, T. R., R. Gillis, S. Marx, C. Brown, and B. H. Iglewski. 2001. Quorum-sensing genes in Pseudomonas aeruginosa biofilms: their role and expression patterns. Appl. Environ. Microbiol. 67:1865-1873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Deziel, E., Y. Comeau, and R. Villemur. 2001. Initiation of biofilm formation by Pseudomonas aeruginosa 57RP correlates with emergence of hyperpiliated and highly adherent phenotypic variants deficient in swimming, swarming, and twitching motilities. J. Bacteriol. 183:1195-1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Egan, S., S. James, C. Holmstrom, and S. Kjelleberg. 2002. Correlation between pigmentation and antifouling compounds produced by Pseudoalteromonas tunicata. Environ. Microbiol. 4:433-442. [DOI] [PubMed] [Google Scholar]
- 14.Egan, S., S. James, and S. Kjelleberg. 2002. Identification and characterization of a putative transcriptional regulator controlling the expression of fouling inhibitors in Pseudoalteromonas tunicata. Appl. Environ. Microbiol. 68:372-378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Egan, S., T. Thomas, C. Holmstrom, and S. Kjelleberg. 2000. Phylogenetic relationship and antifouling activity of bacterial epiphytes from the marine alga Ulva lactuca. Environ. Microbiol. 2:343-347. [DOI] [PubMed] [Google Scholar]
- 16.Fernandez, M., and J. Sanchez. 2002. Nuclease activities and cell death processes associated with the development of surface cultures of Streptomyces antibioticus ETH 7451. Microbiology 148:405-412. [DOI] [PubMed] [Google Scholar]
- 17.Fux, C. A., S. Wilson, and P. Stoodley. 2004. Detachment characteristics and oxacillin resistance of Staphylococcus aureus biofilm emboli in an in vitro catheter infection model. J. Bacteriol. 186:4486-4491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Geesey, G. G., W. T. Richardson, H. G. Yeomans, R. T. Irvin, and J. W. Costerton. 1977. Microscopic examination of natural sessile bacterial populations from an alpine stream. Can. J. Microbiol. 23:1733-1736. [DOI] [PubMed] [Google Scholar]
- 19.Gjermansen, M., P. Ragas, C. Sternberg, S. Molin, and T. Tolker-Nielsen. 2005. Characterization of starvation-induced dispersion in Pseudomonas putida biofilms. Environ. Microbiol. 7:894-906. [DOI] [PubMed] [Google Scholar]
- 20.Gonzalez-Pastor, J. E., E. C. Hobbs, and R. Losick. 2003. Cannibalism by sporulating bacteria. Science 301:510-513. [DOI] [PubMed] [Google Scholar]
- 21.Haussler, S. 2004. Biofilm formation by the small colony variant phenotype of Pseudomonas aeruginosa. Environ. Microbiol. 6:546-551. [DOI] [PubMed] [Google Scholar]
- 22.Heydorn, A., A. T. Nielsen, M. Hentzer, C. Sternberg, M. Givskov, B. K. Ersboll, and S. Molin. 2000. Quantification of biofilm structures by the novel computer program COMSTAT. Microbiology 146:2395-2407. [DOI] [PubMed] [Google Scholar]
- 23.Holmstrom, C., S. James, B. A. Neilan, D. C. White, and S. Kjelleberg. 1998. Pseudoalteromonas tunicata sp. nov., a bacterium that produces antifouling agents. Int. J. Syst. Bacteriol. 48:1205-1212. [DOI] [PubMed] [Google Scholar]
- 24.Holmstrom, C., and S. Kjelleberg. 1999. Marine Pseudoalteromonas species are associated with higher organisms and produce biologically active extracellular agents. FEMS Microbiol. Ecol. 30:285-293. [DOI] [PubMed] [Google Scholar]
- 25.Hunt, S. M., E. M. Werner, B. Huang, M. A. Hamilton, and P. S. Stewart. 2004. Hypothesis for the role of nutrient starvation in biofilm detachment. Appl. Environ. Microbiol. 70:7418-7425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jackson, D. W., K. Suzuki, L. Oakford, J. W. Simecka, M. E. Hart, and T. Romeo. 2002. Biofilm formation and dispersal under the influence of the global regulator CsrA of Escherichia coli. J. Bacteriol. 184:290-301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.James, S. G., C. Holmstrom, and S. Kjelleberg. 1996. Purification and characterization of a novel antibacterial protein from the marine bacterium D2. Appl. Environ. Microbiol. 62:2783-2788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kaplan, J. B., M. F. Meyenhofer, and D. H. Fine. 2003. Biofilm growth and detachment of Actinobacillus actinomycetemcomitans. J. Bacteriol. 185:1399-1404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kaplan, J. B., C. Ragunath, N. Ramasubbu, and D. H. Fine. 2003. Detachment of Actinobacillus actinomycetemcomitans biofilm cells by an endogenous beta-hexosaminidase activity. J. Bacteriol. 185:4693-4698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kerr, C. J., K. S. Osborn, A. H. Rickard, G. D. Robson, and P. S. Handley. 2003. Biofilms in water distribution systems, p. 757-776. In D. Mara and N. J. Horan (ed.), Water and wastewater engineering. Academic Press, London, United Kingdom.
- 31.Kirisits, M. J., L. Prost, M. Starkey, and M. R. Parsek. 2005. Characterization of colony morphology variants isolated from Pseudomonas aeruginosa biofilms. Appl. Environ. Microbiol. 71:4809-4821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kjelleberg, S., and S. Molin. 2002. Is there a role for quorum sensing signals in bacterial biofilms? Curr. Opin. Microbiol. 5:254-258. [DOI] [PubMed] [Google Scholar]
- 33.Lee, S. F., Y. H. Li, and G. H. Bowden. 1996. Detachment of Streptococcus mutans biofilm cells by an endogenous enzymatic activity. Infect. Immun. 64:1035-1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mai-Prochnow, A., F. Evans, D. Dalisay-Saludes, S. Stelzer, S. Egan, S. James, J. S. Webb, and S. Kjelleberg. 2004. Biofilm development and cell death in the marine bacterium Pseudoalteromonas tunicata. Appl. Environ. Microbiol. 70:3232-3238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Marden, P., A. Tunlid, K. Malmcrona-Friberg, G. Odham, and S. Kjelleberg. 1985. Physiological and morphological changes during short term starvation of marine bacterial isolates. Arch. Microbiol. 142:326-332. [Google Scholar]
- 36.Marshall, D. J., T. F. Bolton, and M. J. Keough. 2003. Offspring size affects the post-metamorphic performance of a colonial marine invertebrate. Ecology 84:3131-3137. [Google Scholar]
- 37.Marshall, D. J., and M. J. Keough. 2003. Variation in the dispersal potential of non-feeding larvae: the desperate larva hypothesis and larval size. Marine Ecol. Prog. Ser. 255:145-153. [Google Scholar]
- 38.Marsollier, L., T. Stinear, J. Aubry, J. P. Saint Andre, R. Robert, P. Legras, A. L. Manceau, C. Audrain, S. Bourdon, H. Kouakou, and B. Carbonnelle. 2004. Aquatic plants stimulate the growth of and biofilm formation by Mycobacterium ulcerans in axenic culture and harbor these bacteria in the environment. Appl. Environ. Microbiol. 70:1097-1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McCann, K. S. 2000. The diversity-stability debate. Nature 405:228-233. [DOI] [PubMed] [Google Scholar]
- 40.Mendez, C., A. F. Brana, M. B. Manzanal, and C. Hardisson. 1985. Role of substrate mycelium in colony development in Streptomyces. Can. J. Microbiol. 31:446-450. [DOI] [PubMed] [Google Scholar]
- 41.Miguelez, E. M., C. Hardisson, and M. B. Manzanal. 1999. Hyphal death during colony development in Streptomyces antibioticus: morphological evidence for the existence of a process of cell deletion in a multicellular prokaryote. J. Cell Biol. 145:515-525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Moller, S., C. Sternberg, J. B. Andersen, B. B. Christensen, J. L. Ramos, M. Givskov, and S. Molin. 1998. In situ gene expression in mixed-culture biofilms: evidence of metabolic interactions between community members. Appl. Environ. Microbiol. 64:721-732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Monk, I. R., G. M. Cook, B. C. Monk, and P. J. Bremer. 2004. Morphotypic conversion in Listeria monocytogenes biofilm formation: biological significance of rough colony isolates. Appl. Environ. Microbiol. 70:6686-6694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Moran, A. L., and R. B. Emlet. 2001. Offspring size and performance in variable environments: field studies on a marine snail. Ecology 82:1597-1612. [Google Scholar]
- 45.Neidhardt, F. C., P. L. Bloch, and D. F. Smith. 1974. Culture medium for enterobacteria. J. Bacteriol. 119:736-747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Oliver, A., R. Canton, P. Campo, F. Baquero, and J. Blazquez. 2000. High frequency of hypermutable Pseudomonas aeruginosa in cystic fibrosis lung infection. Science 288:1251-1254. [DOI] [PubMed] [Google Scholar]
- 47.O'Toole, G., H. B. Kaplan, and R. Kolter. 2000. Biofilm formation as microbial development. Annu. Rev. Microbiol. 54:49-79. [DOI] [PubMed] [Google Scholar]
- 48.Resch, A., B. Fehrenbacher, K. Eisele, M. Schaller, and F. Gotz. 2005. Phage release from biofilm and planktonic Staphylococcus aureus cells. FEMS Microbiol. Lett. 252:89-96. [DOI] [PubMed] [Google Scholar]
- 49.Rice, S. A., K. S. Koh, S. Y. Queck, M. Labbate, K. W. Lam, and S. Kjelleberg. 2005. Biofilm formation and sloughing in Serratia marcescens are controlled by quorum sensing and nutrient cues. J. Bacteriol. 187:3477-3485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rogers, J., A. B. Dowsett, P. J. Dennis, J. V. Lee, and C. W. Keevil. 1994. Influence of temperature and plumbing material selection on biofilm formation and growth of Legionella pneumophila in a model potable water system containing complex microbial flora. Appl. Environ. Microbiol. 60:1585-1592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sadowska, B., A. Bonar, C. von Eiff, R. A. Proctor, M. Chmiela, W. Rudnicka, and B. Rozalska. 2002. Characteristics of Staphylococcus aureus, isolated from airways of cystic fibrosis patients, and their small colony variants. FEMS Immunol. Med. Microbiol. 32:191-197. [DOI] [PubMed] [Google Scholar]
- 52.Sauer, K., A. K. Camper, G. D. Ehrlich, J. W. Costerton, and D. G. Davies. 2002. Pseudomonas aeruginosa displays multiple phenotypes during development as a biofilm. J. Bacteriol. 184:1140-1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sauer, K., M. C. Cullen, A. H. Rickard, L. A. Zeef, D. G. Davies, and P. Gilbert. 2004. Characterization of nutrient-induced dispersion in Pseudomonas aeruginosa PAO1 biofilm. J. Bacteriol. 186:7312-7326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Singh, P. K., A. L. Schaefer, M. R. Parsek, T. O. Moninger, M. J. Welsh, and E. P. Greenberg. 2000. Quorum-sensing signals indicate that cystic fibrosis lungs are infected with bacterial biofilms. Nature 407:762-764. [DOI] [PubMed] [Google Scholar]
- 55.Skovhus, T. L., N. B. Ramsing, C. Holmstrom, S. Kjelleberg, and I. Dahllof. 2004. Real-time quantitative PCR for assessment of abundance of Pseudoalteromonas species in marine samples. Appl. Environ. Microbiol. 70:2373-2382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Southey-Pillig, C. J., D. G. Davies, and K. Sauer. 2005. Characterization of temporal protein production in Pseudomonas aeruginosa biofilms. J. Bacteriol. 187:8114-8126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Stoodley, P., L. Hall-Stoodley, J. D. Boyle, F. Jorgensen, and H. M. Lappin-Scott. 2000. Environmental and genetic factors influencing biofilm structure. In D. G. Allison, P. Gilbert, H. M. Lappin-Scott, and M. Wilson (ed.), Community structure and co-operation in biofilms. Cambridge University Press, Cambridge, United Kingdom.
- 58.Sutherland, I. W. 2001. The biofilm matrix—an immobilized but dynamic microbial environment. Trends Microbiol. 9:222-227. [DOI] [PubMed] [Google Scholar]
- 59.van der Wende, E., W. G. Characklis, and D. B. Smith. 1989. Biofilms and bacterial drinking water quality. Water Res. 23:1313-1322. [Google Scholar]
- 60.Webb, J. S., M. Givskov, and S. Kjelleberg. 2003. Bacterial biofilms: prokaryotic adventures in multicellularity. Curr. Opin. Microbiol. 6:578-585. [DOI] [PubMed] [Google Scholar]
- 61.Webb, J. S., M. Lau, and S. Kjelleberg. 2004. Bacteriophage and phenotypic variation in Pseudomonas aeruginosa biofilm development. J. Bacteriol. 186:8066-8073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Webb, J. S., L. S. Thompson, S. James, T. Charlton, T. Tolker-Nielsen, B. Koch, M. Givskov, and S. Kjelleberg. 2003. Cell death in Pseudomonas aeruginosa biofilm development. J. Bacteriol. 185:4585-4592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wimpenny, J. W. T., and R. Colasanti. 1997. A unifying hypothesis for the structure of microbial biofilms based on cellular automaton models. FEMS Microbiol. Ecol. 22:1-16. [Google Scholar]
- 64.Wireman, J. W., and M. Dworkin. 1977. Developmentally induced autolysis during fruiting body formation by Myxococcus xanthus. J. Bacteriol. 129:798-802. [DOI] [PMC free article] [PubMed] [Google Scholar]