Abstract
The possible existence of endemism among microorganisms resulting from and preserved by geographic isolation is one of the most controversial topics in microbial ecology. We isolated 31 strains of “Spumella-like” flagellates from remote sampling sites from all continents, including Antarctica. These and another 23 isolates from a former study were characterized morphologically and by small-subunit rRNA gene sequence analysis and tested for the maximum temperature tolerance. Only a minority of the Spumella morpho- and phylotypes from the geographically isolated Antarctic continent follow the worldwide trend of a linear correlation between ambient (air) temperature during strain isolation and heat tolerance of the isolates. A high percentage of the Antarctic isolates, but none of the isolates from locations on all other continents, were obligate psychrophilic, although some of the latter were isolated at low ambient temperatures. The drastic deviation of Antarctic representatives of Spumella from the global trend of temperature adaptation of this morphospecies provides strong evidence for geographic transport restriction of a microorganism; i.e., Antarctic protistan communities are less influenced by transport of protists to and from the Antarctic continent than by local adaptation, a subtle form of endemism.
One hot debate in microbial ecology deals with the geographic restriction of protistan taxa (8, 10-12, 22, 29). Some researchers believe that free-living protistan species are essentially ubiquitous and grow wherever suitable habitats exist. They assume that the microbial community found in any given habitat is a function of habitat properties only and not of historical factors, since there is an ubiquitous “seed bank.” They further argue that geographic barriers to protists do not exist and consequently cannot contribute to evolutionary diversification (10, 12-15).
The basic assumption that microorganisms are easily dispersed across the physical and geographic barriers that halt the migrations of larger animals and plants (12) is, however, controversial and relies largely on the observed ubiquity of morphospecies. It has been challenged by other researchers, who believe that protists have a biogeography that has been shaped by geographic barriers (8, 30). The discussion has initiated much recent research into the existence of endemic species (10, 16, 28, 38). However, even the definition of a species is controversial, and the low resolution of the morphospecies concept for protists appears to be an obstacle to the settling of the debate (32).
Fortunately, increasing evidence for phylogenetic and ecophysiological differentiation below the morphospecies level (4, 21, 22, 26, 29, 38) can theoretically be used to solve the question of possible endemism among protists. If there were no geographic barriers to protists, correlations between the habitat temperature, as a factor selecting certain strains from the seed bank, and the temperature tolerance of these strains should be independent of the geographic location. To critically test the temperature adaptation of a flagellate morphospecies, we analyzed 54 strains for their phylogenetic relationship and maximum growth temperatures, the latter as a characteristic that is easily measurable and likely subject to evolutionary adaptation to local conditions. Particular attention was paid to isolates from the geographically most isolated sites, on the Antarctic continent.
MATERIALS AND METHODS
Media, isolation of strains, and cultivation conditions.
The standard medium for the maintenance of strains and for the isolation process, except for the first step, was an artificial inorganic basal medium (NSY basal inorganic medium [18]). The ultramicrobacterial (<0.1-μm3) strain MWH-NR1 (Betaproteobacteria; Polynucleobacter group) and the bacterial strain Listonella pelagia CB5 (both available from M. Hahn [17]), representatives of typical free-living aquatic bacteria, were used as food for isolating the flagellates.
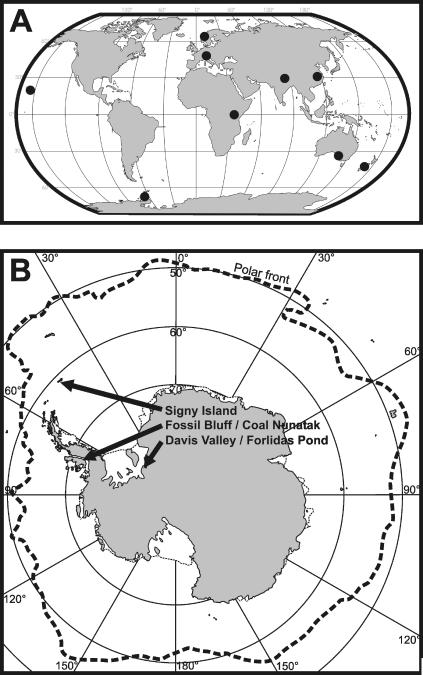
Samples originating from terrestrial and aquatic habitats in polar, temperate, subtropical, and tropical regions (Fig. 1A and B) were collected. Samples were transported to the laboratory in sealed tubes and processed immediately upon arrival. All treatments after sampling were carried out under aseptic conditions. Flagellates were isolated by serial dilution using either sterile filtrated water from the sampling site or artificial media following an acclimatization approach (4). The flagellates were counted using a Sedgewick-Rafter chamber, and a subsample was diluted to a final flagellate abundance of 0.5 to 1 flagellate ml−1 and subsequently transferred to 24-well cell culture plates. Wells were supplemented with food bacteria at a concentration of 3 × 106 to 5 × 106 bacteria ml−1. Wells were checked every second day for a period of at least 2 weeks for positive growth under the microscope, using a total magnification of ×200. When flagellate growth was detected, the medium was transferred to a 50-ml Erlenmeyer flask containing inorganic NSY medium and fresh food bacteria. After 2 to 6 days the subsamples were further diluted to final concentrations of 0.05, 0.1, 0.2, and 0.4 flagellate ml−1 and supplemented with the bacterial strain MWH-Mo1 or CB5 at concentrations of 15 × 106 to 25 × 106 bacteria ml−1. Each of these dilutions was transferred to wells of sterile 24-well cell culture plates (1 ml per well) and incubated at 15 to 22°C, depending on the origin of the isolates. Screening of the wells for the growth of flagellates was again performed by direct microscopic investigation every second day. Finally, flagellates were transferred to an Erlenmeyer flask containing fresh medium and bacteria. This procedure was repeated until pure cultures were established, but at least four times. Pure cultures were acclimatized to 16°C and transferred to permanent culture with NSY medium supplemented with wheat grains. Flagellates in permanent culture were grown at 16°C at low light and transferred once a month to fresh medium. Flagellate strains are available from the authors upon request.
FIG. 1.
Sampling sites. Samples originated from different locations in Europe, Asia, Australia, New Zealand, Hawaii, and Antarctica (A). Each dot represents a sampling area comprising several remote sampling sites. For instance, the Antarctic isolates originate from habitats in three different sampling areas (B). For details, see Table S1 in the supplemental material.
Morphological and phylogenetic analyses.
Cells were checked for the occurrence of chloroplasts. In addition, cells were checked for autofluorescence as follows. Cells were fixed with formaldehyde (final concentration, 2%) and stained with DAPI (4′,6′-diamidino-2-phenylindole) (final concentration, 10 μg ml−1) for 30 min. The cells were then filtered onto a black Nuclepore 0.2-μm filter backed by a 0.45-μm cellulose nitrate filter and examined under an epifluorescence microscope, using UV and blue light excitation for DAPI and for chlorophyll autofluorescence, respectively. Representative strains were, in addition, checked by scanning electron microscopy (SEM) for tripartite hairs on the long flagellum and for occurrence of scales. Briefly, cells fixed with a mixture of Lugol's solution and formaldehyde were allowed to sediment onto glass coverslips covered with poly-l-lysine solution (0.1%, wt/vol). The coverslips were then recovered from the chambers and rinsed in distilled water or sodium cacodylate buffer. Subsequently, the slides were fixed for 30 min in an osmium tetroxide solution (2% final concentration), rinsed again, and subsequently dehydrated using increasingly concentrated ethanol baths and a final wash in hexamethyldisilyazane. Finally, the slides were glued to an SEM stub with a thin layer of Araldite glue, metal coated, and inspected. The phylogenetic affiliation of strains based on small-subunit (SSU) rRNA gene sequences was specified as described in a previous study (4). PCR products were either directly sequenced or used for subsequent cloning into the vector pGEM-T Easy (Promega). Sequencing reactions were performed with an ABI Prism BigDye Terminator version 3.0 Ready Reaction cycle sequencing kit (Applied Biosciences) and an ABI PRISM model 3100 automated sequencer. Sequences were submitted to the BLAST search program of the National Center for Biotechnology Information (NCBI) to find closely related sequences. Sequences were aligned using the “CLUSTAL W” option (34) in the BioEdit 5.0.9 sequence analysis software (19). Where necessary, alignments were subsequently manually processed and corrected. Positions of unclear homology were excluded from further phylogenetic analysis. The result was a final alignment of 1,563 positions. The TREECON 1.3b software package was used to calculate distance matrices by the Kimura algorithm (20) and to generate phylogenetic trees by the neighbor-joining method (36). Parsimony trees were calculated using the program package PHYLIP (version 3.5; J. Felsenstein, Department of Genetics, University of Washington, Seattle). One hundred bootstrapped replicate resampling data points were generated with SEQBOOT (PHYLIP).
Maximum temperature tolerance.
The isolated colorless chrysophytes were characterized regarding their temperature tolerance. During permanent culture, all strains were acclimatized to the same conditions, i.e., NSY inorganic basal medium at 16°C and pH 7.8, for at least 3 months. All experiments were run in triplicate in 4 ml in 12-well tissue culture plates with the bacterial strain Listonella pelagia CB5 as the food source. The strains were transferred to the experimental conditions, and growth was checked every 2 to 3 days by inverted light microscopy at a magnification of ×200. The experiments ran usually for 3 days but up to 8 days until growth was observed. If no growth was observed after 8 days, the treatment was accepted not to support growth of the tested strain. If growth was observed, an aliquot was transferred to fresh medium and food at the next-higher temperature treatment in steps of 1 to 2°C (the temperatures tested were 17.3, 18.0, 19.2, 20.8, 23.0, 24.4, 25.2, 26.7, 28.0, 28.7, 30.7, 31.7, 32.0, 33.6, 34.6, 35.7, 36.4, 37.3, and 37.5°C). This general setup allowed for a stepwise acclimatization of the flagellate strains to the increasing temperatures tested. We generally followed this acclimatization approach, even though successful direct transfer to different temperatures provided evidence that acclimatization hardly affected the tolerance limits. In the context of this work, acclimatization is a physiological or behavioral adjustment to changes in the environment and adaptation is a trait that has evolved over a period of time and is based on a molecular mutation. All experiments were run in triplicate. Using this general strategy, the upper temperature tolerance limit was determined.
Temperature data for the habitats during sampling were not available for all habitats. Therefore, we used the mean monthly air temperatures in the respective month of sampling based on the data set for the past 10 years of the nearest weather station data provided by the Goddard Institute for Space Studies (http://data.giss.nasa.gov/gistemp/station_data/). For some climate stations recent data were not available, and in these cases temperatures were estimated by regression of the available data sets. Where necessary, temperature data were corrected by assuming a mean decrease in air temperature with altitude of 0.7°C per 100 m (temperature decline usually varies between 0.6 and 0.8°C per 100 m [see, e.g., reference 31]).
Nucleotide sequence accession numbers.
The almost-full-length 18S rRNA gene sequences determined in this study have been deposited in the NCBI database under accession numbers DQ388538 to DQ388568.
RESULTS
Morphological and 18S rRNA gene analyses.
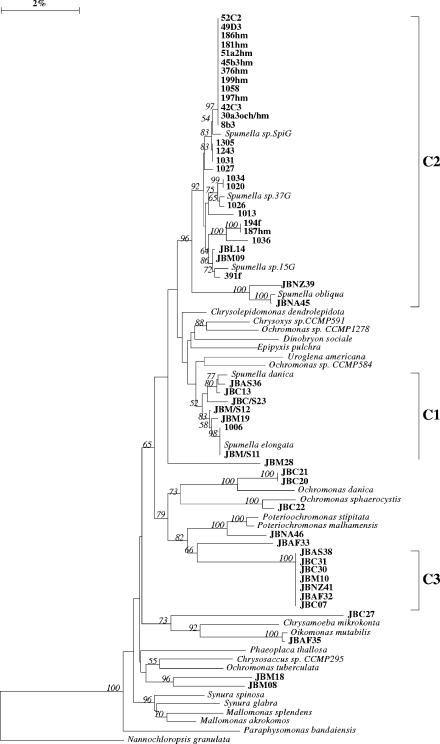
We isolated 31 colorless chrysophyte flagellates identified as Spumella spp. These isolates and, in addition, 23 isolates from our culture collection (4) were included in this study. The Spumella cells were colorless and solitary and had a spherical to ellipsoid cell form. From the front end emerged two flagella, both visible in the light microscope. SEM investigations proved that the long one bears tripartite hairs. The cells either swim free in the medium or attach to the substrate with the posterior end. The cells feed on small particles, specifically bacteria, and the general sequence of feeding follows that described by Boenigk and Arndt (3). The cells were between 4 and 8 μm in diameter. Despite a detailed morphological investigation of our Spumella strains, a classification to one of the described taxa was not possible due to the poor quality of the existing taxon descriptions (2). We isolated Spumella spp. from all continents from contrasting freshwater and soil sites (Fig. 1). Based on morphology, the investigated species therefore seem to be ubiquitous, i.e., present in different habitat types and distributed worldwide. Both neighbor-joining and parsimony 18S rRNA gene trees showed that most of our Spumella isolates were affiliated with the clusters C1, C2, and C3 as described previously (4) (Fig. 2; see Fig. S1 in the supplemental material). As the phylogenetic analysis indicates polyphyly of Spumella spp., we further use the term “Spumella-like” flagellates.
FIG. 2.
Neighbor-joining tree showing the affiliation of SSU rRNA gene sequences from Spumella isolates with the Chrysophyceae sensu stricto. The numbers at the nodes of the tree indicate the percentage of bootstrap values for each node out of 100 bootstrap resamplings (values above 50 are shown). The scale bar indicates 2% estimated sequence divergence.
Ecophysiological differentiation and indications for geographic restriction.
The “Spumella-like” flagellates differed significantly regarding their maximum tolerated growth temperature (by analysis of variance, P < 0.001) (Fig. 2), ranging between 17.3°C for a strain originating from Heywood Lake, Signy Island, Maritime Antarctic, and 34.6°C for several strains from Africa and Asia (see Table S1 in the supplemental material). Except for one strain originating from Antarctica and tolerating 24.4°C, all strains affiliated with the Spumella subclusters C1 and C3 tolerated maximum temperatures of between 28 and 34.6°C and of between 32 and 34.6°C, respectively. Strains from subcluster C2 were split between the Antarctic strains, tolerating between 17.3 and 28°C, and strains from other regions, which tolerated between 30.7 and 32°C (Fig. 2; see Fig. S1 and Table S1 in the supplemental material).
The temperature tolerance of the isolates was correlated with the mean monthly and mean annual air temperatures at the isolation sites (Spearman rank order correlations, r = 0.857 [P < 0.001] and r = 0.867 [P < 0.001], respectively). Similar correlation with monthly and annual temperatures at the sampling sites indicates little seasonality.
The habitat type had no effect on this correlation (for soil versus freshwater, by analysis of covariance [ANCOVA], P = 0.624).
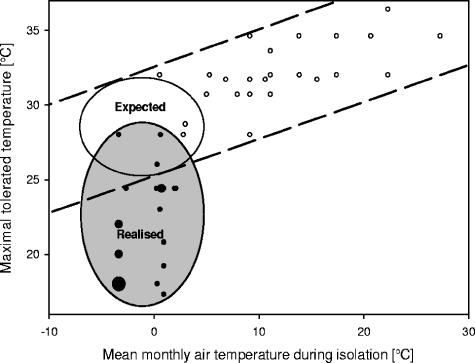
The temperature tolerance of the isolates, excluding those from Antarctica, rose linearly with ambient air temperature (by linear regression, adjusted r2 = 0.36 [P < 0.001]) (Fig. 3). Most of the strains from Antarctic samples deviated significantly from the global trend in that they were more strongly adapted to cold conditions (Fig. 3), including several psychrophilic strains (temperature maxima of <20°C). This was significant when assuming either a linear or a logarithmic model (for a linear model, by ANCOVA, P = 0.003; for log-transformed data, by ANCOVA, P < 0.001). The test for the two different model assumptions was done because regression analysis yielded similar regression coefficients for both models (r2 = 0.685; log (X + 5) transformed, r2 = 0.729).
FIG. 3.
Geographic pattern of temperature tolerance of Spumella-like flagellates. Maximum growth temperatures differ between strains but are generally within certain limits that relate to environmental temperature. Strains originating from Antarctica are represented by black symbols; those from other continents are represented by white symbols. For the latter there is a global linear correlation of increasing temperature tolerance with increasing environmental temperature (dashed lines). Antarctic strains deviate from this trend; i.e., the realized temperature adaptation (gray oval) is stronger than expected from the global trend (white oval). The size of the symbol indicates the number of strains for the respective condition; i.e., small symbols represent one strain, medium-sized symbols represent two or three strains; and large symbols represent five strains.
DISCUSSION
Strains affiliated with the same morphotype differ considerably with respect to ecophysiology.
Morphological analysis of heterotrophic protistan communities from several marine and freshwater sites around Antarctica yielded taxa of which most had been previously reported from geographic locations elsewhere (6, 35), thus supporting the view that species may be worldwide distributed. Counter to this view of limited endemism are observations that the Antarctic region is more isolated than other parts of the world and that there has been environmental selection for specific adaptive strategies over a period of several million years (37). The morphological approach, indicating a cosmopolitan and ubiquitous distribution of most protists (12), may indeed be inadequate, as molecular studies provide evidence for considerable microdiversity and a certain geographic isolation of organisms in Antarctica. For protists (23), cyanobacteria (33), and heterotrophic bacteria (37), molecular data indicate a high level of geographic isolation. This study provides evidence for ecophysiological differentiation below the level of morphospecies but also below the level of SSU rRNA clusters. Similarly, a high functional diversity has been demonstrated for several flagellate taxa, specifically for Oxyrris marina (26), Neobodo designis (21, 38), and Spumella (5). These studies agree that ecophysiological parameters vary within a morphospecies. So far, the available studies do not support a clear correlation between ecophysiological and molecular differences, even though several studies indicate a habitat specificity on the level of SSU rRNA clusters (4, 21, 26, 38). All these studies provide indications that morphological characters may be insufficient for a valid taxonomic classification.
Based on the current knowledge a solution of the species problem in protists seems to still be a distant prospect. In the meantime, we suggest that all available data be considered, including morphology, molecular data, and ecophysiological data, to define protistan species. Our data indicate that strains identical in the SSU rRNA gene sequence seem to have similar ecophysiological characteristics as well. Ecophysiological variation, however, seems generally to be high even between closely related flagellate strains (5; this study). Even slight differences in the SSU rRNA gene sequence may already be affiliated with very different ecophysiological adaptations. Further, we have to keep in mind that eventually any species concept, i.e., a classification of organisms in distinct units, may be inappropriate or at least problematic to describe the continuous transitions that we observe in the ecophysiology of protistan strains and taxa.
Antarctic strains are cold adapted.
Our data clearly indicate an adaptation of Antarctic strains to the cold environment, even though the tolerated temperatures are far above the realized temperatures in the respective environments. We followed a conservative protocol; i.e., all strains were acclimatized to 16°C before the experiments. The deviation of the Antarctic strains from the global trend is therefore likely to be even stronger than observed in our experiment. Further, the relatively high tolerated temperatures of the cold-adapted flagellates, i.e., between 17.3 and 28°C, correspond to theoretical considerations: low-temperature adaptation of enzymes is regarded as an ongoing process, and optimal adaptation is therefore not to be expected (9). Cold adaptation of Antarctic chrysophyte flagellates has already been demonstrated for Paraphysomonas (7) and is further supported by observations of community growth of Antarctic flagellates at low temperature (25). In contrast, based on low realized growth rates of the Antarctic heterotrophic nanoflagellate community in Crooked Lake at 2°C, Laybourn-Parry et al. (24) concluded that these flagellates were not adapted. Those authors, however, did not exclude predation in their experiments, and the bacterial food concentration was below 5 × 105 bacteria ml−1 with a mean bacterial cell volume of around 0.1 μm3. Under such conditions our isolates, originating from warmer habitats, would even die back (5; J. Boenigk et al. unpublished data), and we therefore do not see a conflict with the earlier study. For the investigated strains, temperature adaptation seems not to be linked to habitat characteristics, and SSU rRNA data may not provide sufficient resolution to separate ecophysiologically different clusters. Even flagellate strains identical in SSU rRNA sequence (5) and flagellate strains originating from the same habitat (this study) may differ considerably in their ecophysiology. We therefore advise caution in the ecological interpretation of experiments based on a single strain.
Geographic barriers do matter for the distribution of protists.
The west wind zone and the mainly wind-driven Antarctic circumpolar current, i.e., the strongest oceanic current (1), are the main reasons for the (bio-)geographic isolation of Antarctica and its stable cold climate. Thus, if “microbial endemism” is possible at all, then Antarctica would be a promising place to find such organisms (37). In fact, the drastic deviation of the temperature adaptation of Antarctic Spumella morphospecies from the global trend provides strong evidence for geographic transport restriction of evolutionary and, consequently, biogeographic significance. Assuming unlimited exchange of protists, one would expect that the global trend of a steady decrease in temperature tolerance with environmental temperature includes the Antarctic continent. This does not question the possibility that protistan cells are transported by air between geographic regions (37). Intercontinental transport of protists, as proposed by Finlay and Fenchel (10, 12), is very likely (cf. references 2 and 4) and is supported by the observation of significant air travel of spores and pollen to Antarctica (27). Further, the phylogenetic similarity of isolates from Antarctic and non-Antarctic sampling sites indicates recent (in evolutionary time scales) exchange. Our data show, however, that protistan transport to Antarctica is sufficiently restricted to allow the local protistan population to adapt (not only acclimatize) to local environmental conditions and thus to build up biogeographically restricted populations. The predominance of autochthonous strains from Antarctica that are identifiable from their deviation from the global trend of temperature adaptation (Fig. 2) demonstrates that successful colonization of Antarctic habitats by allochthonous strains is rare.
Based on our findings, we expect subtle cases of protistan endemism to exist also in other isolated habitats shielded by geographic barriers, as proposed by those critical of the ubiquity theorem (8, 16, 29). We assume, however, that the resolution of current taxonomic methods, including molecular analyses, may overlook endemic ecophysiological traits as well as variance at the whole-genome level. Applying finer taxonomic resolution, we may eventually find evidence for other geographic barriers to microbes. The existence of endemic morphospecies appears unlikely, since rates of global transport of microorganisms, although restricted, are too much higher than evolutionary speciation into morphologically distinct organisms. Whether microbes can be endemic or have a biogeography depends on whether we continue to restrict these terms to evolutionary distances separating species or higher taxa. It should be noted, however, that there are less permissive and durable barriers, such as geological formations or ice caps, that may give rise to geographic speciation of microbes as is known for plants and animals.
Supplementary Material
Acknowledgments
Parts of this work were funded by the Austrian Science Fund (P 18767) and by the Natural Environment Research Council through the British Antarctic Survey Terrestrial and Freshwater Biodiversity Programme.
Footnotes
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Barker, P. F., and E. Thomas. 2004. Origin, signature and palaeoclimatic influence of the Antarctic Circumpolar Current. Earth-Sci. Rev. 66:143-162. [Google Scholar]
- 2.Boenigk, J. 2005. Some remarks on the strain specificty and general pattern in the ecology of Spumella (Chrysophyceae). Nova Hedwigia 128:167-178. [Google Scholar]
- 3.Boenigk, J., and H. Arndt. 2000. Particle handling during interception feeding by four species of heterotrophic nanoflagellates. J. Eukaryot. Microbiol. 47:350-358. [DOI] [PubMed] [Google Scholar]
- 4.Boenigk, J., K. Pfandl, P. Stadler, and A. Chatzinotas. 2005. High diversity of the “Spumella-like” flagellates: an investigation based on the SSU rRNA gene sequences of isolates from habitats located in six different geographic regions. Environ. Microbiol. 7:685-697. [DOI] [PubMed] [Google Scholar]
- 5.Boenigk, J., P. Stadler, A. Wiedlroither, and M. W. Hahn. 2004. Strain-specific differences in the grazing sensitivity of closely related ultramicrobacteria affiliated with the Polynucleobacter cluster. Appl. Environ. Microbiol. 70:5787-5793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Butler, H. G. 1999. Seasonal dynamics of the planktonic microbial community in a maritime antarctic lake undergoing eutrophication. J. Plankton Res. 21:2393-2419. [Google Scholar]
- 7.Choi, J. W., and F. Peters. 1992. Effects of temperature on two psychrophylic ecotypes of a heterotrophic nanoflagellate, Paraphysomonas imperforata. Appl. Environ. Microbiol. 58:593-599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Coleman, A. W. 2002. Microbial eukaryote species. Science 297:337. [DOI] [PubMed] [Google Scholar]
- 9.D'amico, S., P. Claverie, T. Collins, D. Georlette, E. Gratia, A. Hoyoux, M. A. Meuwis, G. Feller, and C. Gerday. 2002. Molecular basis of cold adaptation. Phil. Trans. R. Soc. London B 357:917-924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fenchel, T., and B. J. Finlay. 2004. The ubiquity of small species: patterns of local and global diversity Bioscience 54:777-784.
- 11.Fenchel, T., and B. J. Finlay. 2004. Here and there or everywhere? Response from Fenchel and Finlay. Bioscience 54:884-885. [Google Scholar]
- 12.Finlay, B. J. 2002. Global dispersal of free-living microbial eukaryote species. Science 296:1061-1063. [DOI] [PubMed] [Google Scholar]
- 13.Finlay, B. J., and K. J. Clarke. 1999. Ubiquitous dispersal of microbial species. Nature 400:828. [Google Scholar]
- 14.Finlay, B. J., and K. J. Clarke. 1999. Apparent global ubiquity of species in the protist genus Paraphysomonas. Protist 150:419-430. [DOI] [PubMed] [Google Scholar]
- 15.Finlay, B. J., and T. Fenchel. 2002. Microbial eukaryote species—response. Science 297:337. [DOI] [PubMed] [Google Scholar]
- 16.Foissner, W. 2005. Two new “flagship” ciliates (Protozoa, Ciliophora) from Venezuela: Sleighophrys pustulata and Luporinophrys micelae. Eur. J. Protistol. 41:99-117. [Google Scholar]
- 17.Hahn, M. W. 1997. Experimentelle Untersuchungen zur Interaktion von bakterivoren Nanoflagellaten mit pelagischen Bakterien. Thesis. TU Braunschweig, Braunschweig, Germany.
- 18.Hahn, M. W., H. Lunsdorf, Q. L. Wu, M. Schauer, M. G. Hofle, J. Boenigk, and P. Stadler. 2003. Isolation of novel ultramicrobacteria classified as actinobacteria from five freshwater habitats in Europe and Asia. Appl. Environ. Microbiol. 69:1442-1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hall, T. A. 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. 41:95-98. [Google Scholar]
- 20.Kimura, M. 1980. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 16:11-120. [DOI] [PubMed] [Google Scholar]
- 21.Koch, T. A., and F. Ekelund. 2005. Strains of the heterotrophic flagellate Bodo designis from different environments vary considerably with respect to salinity preference and SSU rRNA gene composition. Protist 156:97-112. [DOI] [PubMed] [Google Scholar]
- 22.Lachance, M.-A. 2004. Here and there or everywhere? Bioscience 54:884. [Google Scholar]
- 23.Lawley, B., S. Ripley, P. Bridge, and P. Convey. 2004. Molecular analysis of geographic patterns of eukaryotic diversity in antarctic soils. Appl. Environ. Microbiol. 70:5963-5972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Laybourn-Parry, J., P. Bayliss, and J. C. Ellisevans. 1995. The dynamics of heterotrophic nanoflagellates and bacterioplankton in a large ultra-oligotrophic Antarctic lake. J. Plankton Res. 17:1835-1850. [Google Scholar]
- 25.Laybourn-Parry, J., J. C. Ellisevans, and H. Butler. 1996. Microbial dynamics during the summer ice-loss phase in maritime Antarctic lakes. J. Plankton Res. 18:495-511. [Google Scholar]
- 26.Lowe, C. D., A. Day, S. J. Kemp, and D. J. S. Montagnes. 2005. There are high levels of functional and genetic diversity in Oxyrrhis marina. J. Eukaryot. Microbiol. 52:250-257. [DOI] [PubMed] [Google Scholar]
- 27.Marshall, M. A. 1996. Biological particles over Antarctica. Nature 383:680. [Google Scholar]
- 28.Massana, R., J. Castresana, V. Balague, L. Guillou, K. Romari, A. Groisillier, K. Valentin, and C. Pedros-Alio. 2004. Phylogenetic and ecological analysis of novel marine stramenopiles. Appl. Environ. Microbiol. 70:3528-3534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nanney, D. L. 2004. No trivial pursuit. Bioscience 54:720-721. [Google Scholar]
- 30.Papke, R. T., and D. M. Ward. 2004. The importance of physical isolation to microbial diversification. FEMS Microb. Ecol. 48:293-303. [DOI] [PubMed] [Google Scholar]
- 31.Roedel, W. 2000. Physik unserer Umwelt: Die Atmosphäre. 3. Aufl. Springer Verlag, Berlin, Germany.
- 32.Schlegel, M., and R. Meisterfeld. 2003. The species problem in protozoa revisited. Eur. J. Protistol. 39:349-355. [Google Scholar]
- 33.Taton, A., S. Grubisic, E. Brambilla, R. De Wit, and A. Wilmotte. 2003. Cyanobacterial diversity in natural and artificial microbial mats of Lake Fryxell (McMurdo Dry Valleys, Antarctica): a morphological and molecular approach. Appl. Environ. Microbiol. 69:5157-5169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Thompson, J. D., T. J. Gibson, F. Plewniak, Jeanmougin. F., and D. G. Higgins. 1997. The Clustal X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acid Res. 24:4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tong, S., N. Vors, and D. J. Patterson. 1997. Heterotrophic flagellates, centrohelid heliozoa and filose amoebae from marine and freshwater sites in the Antarctic. Polar Biol. 18:91-106. [Google Scholar]
- 36.van de Peer, Y., and R. de Wachter. 1994. TREECON for Windows: a software package for the construction and drawing of evolutionary trees for the Microsoft Windows environment. Comput. Appl. Biosci. 10:569-570. [DOI] [PubMed] [Google Scholar]
- 37.Vincent, W. F. 2000. Evolutionary origins of Antarctic microbiota: invasion, selection and endemism. Antarctic Sci. 12:374-385. [Google Scholar]
- 38.von der Heyden, S., and T. Cavalier-Smith. 2005. Culturing and environmental DNA sequencing uncover hidden kinetoplastid biodiversity and a major marine clade within ancestrally freshwater Neobodo designis. Int. J. Syst. Evol. Microbiol. 55:2605-2621. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.