Abstract
Non-Candida albicans Candida species are increasingly being isolated. These species show differences in levels of resistance to antimycotic agents and mortality. Therefore, it is important to be able to correctly identify the causative organism to the species level. Identification of C. dubliniensis in particular remains problematic due to the high degree of phenotypic similarity between this species and C. albicans. The use of amplified fragment length polymorphism (AFLP) analysis as an identification method for medically important Candida species was investigated. Our results show very clear differences among medically important Candida species. Furthermore, when screening a large collection of clinical isolates previously identified on CHROMagar as C. albicans, we found a misidentification rate of 6%. AFLP analysis is universally applicable, and the patterns can easily be stored in a general, accessible database. Therefore, AFLP might prove to be a reliable method for the identification of medically important Candida species.
In the past decade, the number of life-threatening forms of candidiasis increased dramatically (1). The attributable mortality of these infections is as high as 38% (34), whereas crude mortality rates exceed 50% (10, 27, 33). For a long time, Candida albicans was the main cause of invasive fungal infections. However, the number of infections by this species is declining whereas non-albicans Candida species like C. glabrata, C. krusei, and C. parapsilosis are increasingly being isolated. At present, non-albicans Candida species account for approximately 50% of all Candida infections (14).
In cases of candidiasis, it is important to be able to correctly identify the causative organism to the species level. Different species show differences in levels of resistance to antimycotic agents. C. krusei is innately resistant to fluconazole, and C. glabrata is able to acquire resistance to this drug rapidly. Furthermore, C. glabrata infections have been associated with a high mortality (11). A particular problem is formed by the recently recognized species C. dubliniensis. Like C. glabrata, this species is capable of acquiring stable fluconazole resistance rapidly (22, 23). Identification of C. dubliniensis remains difficult, due to the high degree of phenotypic similarity between this species and C. albicans. However, it is known that genotypically there is more variation between the two species (30). Therefore, molecular identification methods may be more reliable than identification methods based on phenotypic characteristics.
Amplified fragment length polymorphism (AFLP) analysis is a relatively new technique which has a discriminatory power that makes it suitable for identification as well as for strain typing (29, 32). In short, in AFLP analysis genomic DNA is digested with two restriction enzymes (e.g., EcoRI and MseI) and double-stranded oligonucleotide adapters are ligated to the fragments. These adapters serve as targets for the primers during PCR amplification. To increase the specificity, it is possible to elongate the primers at their 3′ ends with one to three selective nucleotides. One of the primers is labeled with a fluorescent dye. The fragments are separated and analyzed using software packages like BioNumerics (Applied Maths, Sint-Martens-Latem, Belgium). The advantage of AFLP analysis is that only a limited amount of DNA is needed since the fragments are PCR amplified. Furthermore, since stringent annealing temperatures are used during amplification, the technique is more reproducible and robust than other methods such as randomly amplified polymorphic DNA analysis (13, 29). This paper describes the use of AFLP as an identification method for medically important Candida species, including C. dubliniensis.
MATERIALS AND METHODS
Yeast strains.
The yeast strains and isolates used are listed in Table 1. Reference strains were obtained from the Centraalbureau voor Schimmelcultures (CBS), Utrecht, The Netherlands, and the American Type Culture Collection, Manassas, Va. Clinical isolates were obtained from the European SENTRY collection (Eijkman-Winkler Center, Utrecht, The Netherlands) (26) and from the hematology ward of the VU University Medical Center (VUMC), Amsterdam, The Netherlands. Upon receipt of the SENTRY isolates in the central laboratory (Eijkman-Winkler Center), CHROMagar (CHROMagar, Paris, France) was used to distinguish between C. albicans and non-C. albicans species. Non-C. albicans species were further identified by using the API Candida system (bioMérieux, Marcy-l'Etoile, France). In addition, Vitek YBC cards (bioMérieux) were used when the results obtained by the API Candida system were inconclusive or differed from the identification made by the center where the Candida species was isolated. The isolates from the VUMC were identified using the germ-tube test. The isolates that were negative in this assay were further identified by using Vitek YBC (bioMérieux).
TABLE 1.
Reference strains and clinical isolates used in this study
| Speciesa | Strain or isolate (collection) | Origin | Source |
|---|---|---|---|
| C. albicans | CBS 562 | Uruguay | Skin of man with interdigital mycosis |
| CBS 1905 | Unknown | Man | |
| CBS 1912 | Norway | Sputum of asthma patient | |
| ATCC 90028 | Iowa, United States | Blood | |
| ATCC 90029 | Iowa, United States | Blood | |
| 04A080 (SENTRY) | Paris, France | Blood | |
| 06A309 (SENTRY) | Lille, France | Blood | |
| 07C069 (SENTRY) | Freiburg, Germany | Pneumonia | |
| 08E058 (SENTRY) | Dusseldorf, Germany | Wound, skin, or soft tissue | |
| 10A173 (SENTRY) | Genoa, Italy | Blood | |
| 10C007 (SENTRY) | Genoa, Italy | Pneumonia | |
| 11A134 (SENTRY) | Rome, Italy | Blood | |
| 11C034 (SENTRY) | Rome, Italy | Pneumonia | |
| 12E033 (SENTRY) | Utrecht, The Netherlands | Wound, skin, or soft tissue | |
| 15A020 (SENTRY) | Coimbra, Portugal | Blood | |
| 15A206 (SENTRY) | Coimbra, Portugal | Blood | |
| 15A561 (SENTRY) | Coimbra, Portugal | Blood | |
| 16A232 (SENTRY) | Seville, Spain | Blood | |
| 16A438 (SENTRY) | Seville, Spain | Blood | |
| 16C088 (SENTRY) | Seville, Spain | Pneumonia | |
| 17A381 (SENTRY) | Madrid, Spain | Blood | |
| 19A164 (SENTRY) | Lausanne, Switzerland | Blood | |
| 19A519 (SENTRY) | Lausanne, Switzerland | Blood | |
| 19A567 (SENTRY) | Lausanne, Switzerland | Blood | |
| 19A568 (SENTRY) | Lausanne, Switzerland | Blood | |
| 23D045 (SENTRY) | Ankara, Turkey | Urinary tract | |
| TY727 (VUMC) | Amsterdam, The Netherlands | Oral cavity | |
| TY728 (VUMC) | Amsterdam, The Netherlands | Oral cavity | |
| TY729 (VUMC) | Amsterdam, The Netherlands | Oral cavity | |
| TY732 (VUMC) | Amsterdam, The Netherlands | Feces (human) | |
| C. dubliniensis | CBS 7987 | Dublin, Ireland | Oral cavity of HIV-infected patient |
| CBS 7988 | Melbourne, Australia | Oral cavity of HIV-infected patient | |
| CBS 8500 | Nijmegen, The Netherlands | Blood of 38-year-old woman with chronic myelogenous leukemia | |
| CBS 8501 | Nijmegen, The Netherlands | Child with neutropeny induced by chemotherapy | |
| 02A038 (SENTRY) | Brussels, Belgium | Blood | |
| 05C118 (SENTRY) | Lyon, France | Pneumonia | |
| 05C121 (SENTRY) | Lyon, France | Pneumonia | |
| 18A221 (SENTRY) | Barcelona, Spain | Blood | |
| 20C149 (SENTRY) | London, United Kingdom | Pneumonia | |
| 23A137 (SENTRY) | Ankara, Turkey | Blood | |
| C. glabrata | CBS 138 | Unknown | Feces (human) |
| ATCC 90030 | Iowa, United States | Blood | |
| TY714 (VUMC) | Amsterdam, The Netherlands | Oral cavity | |
| TY715 (VUMC) | Amsterdam, The Netherlands | Feces (human) | |
| TY716 (VUMC) | Amsterdam, The Netherlands | Oral cavity | |
| TY717 (VUMC) | Amsterdam, The Netherlands | Oral cavity | |
| TY718 (VUMC) | Amsterdam, The Netherlands | Oral cavity | |
| TY719 (VUMC) | Amsterdam, The Netherlands | Oral cavity | |
| TY731 (VUMC) | Amsterdam, The Netherlands | Oral cavity | |
| C. guilliermondii | CBS 566 | Unknown | Sputum (human) |
| CBS 2024 | Berlin, Germany | Ulcer on horse | |
| 14A097 (SENTRY) | Cracow, Poland | Blood | |
| C. krusei | CBS 573 | Colombo, Sri Lanka | Sputum of bronchitic convict |
| TY722 (VUMC) | Amsterdam, The Netherlands | Oral cavity | |
| TY723 (VUMC) | Amsterdam, The Netherlands | Oral cavity | |
| TY726 (VUMC) | Amsterdam, The Netherlands | Feces (human) | |
| C. lusitaniae | CBS 4413 | Portugal | Cecum of pig |
| C. parapsilosis | CBS 604 | Puerto Rico | Case of sprue (human) |
| CBS 2195 | Austria | Infected nail of 11-year-old boy | |
| ATCC 90018 | Virginia, United States | Blood | |
| 07A212 (SENTRY) | Freiburg, Germany | Blood | |
| 10A120 (SENTRY) | Genoa, Italy | Blood | |
| 10A311 (SENTRY) | Genoa, Italy | Blood | |
| 14A161 (SENTRY) | Cracow, Poland | Blood | |
| TY735 (VUMC) | Amsterdam, The Netherlands | Oral cavity | |
| TY736 (VUMC) | Amsterdam, The Netherlands | Unknown | |
| C. pseudotropicalis | CBS 607 | Sri Lanka | Bronchitic patient |
| C. tropicalis | CBS 94 | Unknown | Bronchitic patient |
| CBS 2310 | Unknown | Unknown | |
| 11D028 (SENTRY) | Rome, Italy | Urinary tract | |
| TY737 (VUMC) | Amsterdam, The Netherlands | Oral cavity | |
| TY739 (VUMC) | Amsterdam, The Netherlands | Oral cavity |
Identification of SENTRY isolates based on AFLP patterns.
Extraction of DNA.
DNA was extracted from approximately 107 CFU using a DNeasy tissue kit (Qiagen, West Sussex, England) according to the manufacturer's instructions (protocol for isolation of genomic DNA from yeasts). DNA was eluted in 100 μl of elution buffer (buffer AE of the kit) and stored at −20°C.
AFLP. (i) Restriction and ligation of adapters.
The sequences of the adapters and primers used for AFLP analysis are given in Table 2. DNA was extracted from approximately 107 CFU of C. albicans as described above. Five microliters of the DNA samples was added to 5 microliters of restriction-ligation reaction mixture (1× T4 DNA ligase buffer, 0.05 M NaCl, 0.5 μg of bovine serum albumin, 2 pmol of the EcoRI adapter, 20 pmol of the MseI adapter, 80 U of T4 DNA ligase, 1 U of EcoRI, 1 U of MseI) and incubated overnight at 37°C. All enzymes were obtained from New England Biolabs (Beverly, Mass.). The mixture was diluted 1:5 with 0.1× TE (5 mM Tris-HCl [pH 7.5], 1 mM EDTA).
TABLE 2.
Adapter and primer sequences used for AFLP
| Adapter or primers | Sequencea |
|---|---|
| Adapters | |
| EcoRI | 5′-CTCGTAGACTGCGTACC-3′ |
| 3′-CATCTGACGCATGGTTAA-5′ | |
| MseI | 5′-GACGATGAGTCCTGAG-3′ |
| 3′-CTACTCAGGACTCAT-5′ | |
| Primers | |
| EcoRI | 5′-GACTGCGTACCAATTCAC-3′ |
| MseI | 5′-GATGAGTCCTGAGTAAC-3′ |
The bold type represents selective nucleotides (added to the core sequence and used only in the second PCR).
(ii) Preselective and selective PCRs.
Preselective PCR was performed using the core sequences, i.e., primers without extensions. The AFLP primers, core mix, and internal size standard were supplied by Applied Biosystems (Nieuwerkerk aan den IJssel, The Netherlands). Four microliters of diluted restriction-ligation product was added to 15 μl of AFLP amplification core mix, 0.5 μl of the EcoRI core sequence, and 0.5 μl of the MseI core sequence. The mixture was amplified in a GeneAmp PCR System 9700 machine under the following conditions: 2 min at 72°C, followed by 20 cycles of 20 s at 94°C, 30 s at 56°C, and 2 min at 72°C. The PCR product was diluted by adding 25 μl of sterile double-distilled water. In a second PCR more-selective primers were used: EcoRI-AC (labeled with 6-carboxyfluorescein) and MseI-C. The conditions were 2 min at 94°C, followed by 10 cycles consisting of 20 s at 94°C and 30 s at 66°C (with this temperature decreasing 1°C with each succeeding cycle), and a final extension of 2 min at 72°C. This sequence was followed by 25 cycles consisting of 20 s at 94°C, 30 s at 56°C, and 2 min at 72°C and a final incubation of 30 min at 60°C.
(iii) Capillary electrophoresis and data analysis.
The samples were prepared for capillary electrophoresis by adding 2 μl of the selective PCR product to 24 μl of deionized formamide and 1 μl of GeneScan-500 (6-carboxy-X-rhodamine [ROX] labeled) as an internal size standard. They were run on an ABI 310 genetic analyzer for 30 min each. Data were analyzed with the BioNumerics software package, version 2.5 (Applied Maths) by using the Pearson correlation as a similarity coefficient in combination with unweighted pair group method with arithmatic mean cluster analysis. The statistical reliability of the clusters was investigated by using the cophenetic values, which calculate the correlation between the calculated similarities and the dendrogram-derived similarities.
RESULTS AND DISCUSSION
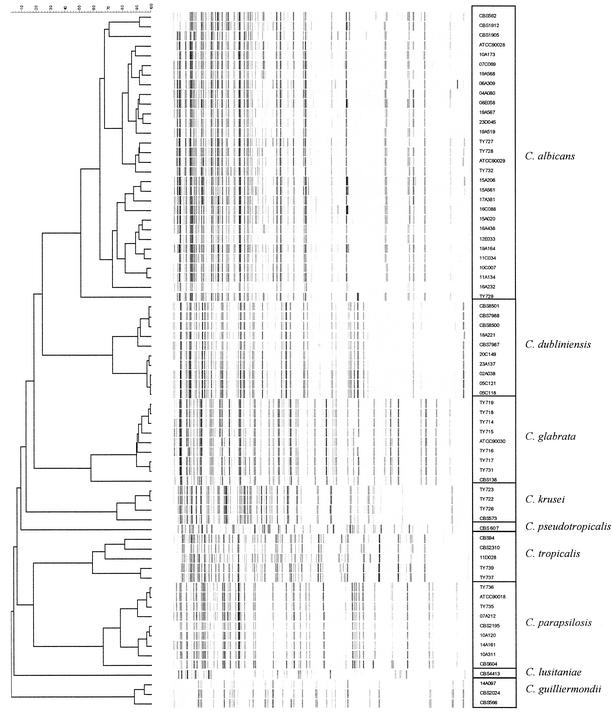
A dendrogram representing all reference strains and clinical isolates is depicted in Fig. 1. The AFLP patterns of the reference strains clearly show that each species forms a distinct cluster. The cophenetic values were 78 for C. albicans, 92 for C. dubliniensis, 99 for C. glabrata, 84 for C. krusei, 98 for C. pseudotropicalis, 85 for C. tropicalis, 91 for C. parapsilosis, 98 for C. lusitaniae, and 94 for C. guilliermondii. These results were highly reproducible.
FIG. 1.
Dendrogram representing all reference strains and clinical isolates (see also Table 1).
The C. albicans isolates show two main clusters. One cluster contains clinical isolates from the VUMC and the SENTRY collection as well as reference strains from the CBS. The other cluster contains only isolates from the SENTRY collection. There is no clear relation between these clusters and the geographical origins or sources of the isolates. North American C. albicans isolates show a three-part division by several typing methods, such as randomly amplified polymorphic DNA analysis, multilocus enzyme electrophoresis, and Southern blot hybridization with the moderately repetitive C. albicans-specific Ca3 probe. In South Africa, an additional cluster besides these three clusters has been found (4, 18, 28). It will be interesting to investigate whether the two AFLP clusters of C. albicans correspond with the North American or South African clusters.
The C. dubliniensis isolates also show two clusters whose isolates have remarkably high similarities (91 and 98%). One cluster contains all reference strains used and one SENTRY clinical isolate; the other cluster is composed of SENTRY isolates only. Using the C. dubliniensis-specific fingerprinting probe Cd25 on a panel of 98 isolates, Gee et al. also recognized two different clusters, one of which contained mainly isolates derived from human immunodeficiency virus (HIV)-infected individuals, while the other cluster contained mainly isolates derived from HIV-negative individuals (9). Strains CBS 7987 and CBS 7988, both part of the same AFLP cluster, were isolated from an HIV-infected individual. However, data on the HIV status of the patients from which the other isolates (CBS 8500, CBS 8501, and SENTRY isolates) were obtained are lacking. Further investigations are necessary to examine whether the AFLP clusters correspond with the Cd25 clusters.
Another noteworthy finding is that all of the AFLP patterns for the C. glabrata isolates are very similar (90% similarity) except for that of the CBS reference strain (58% similarity). This reference strain (CBS 138) was isolated from human feces and was first described in 1917. The fact that all of the other isolates studied were clinical isolates which were isolated fairly recently may account for this difference.
The AFLP patterns of the 18 isolates from the VUMC all corresponded with the results of the phenotypic identification (obtained by using the germ-tube test and Vitek YBC cards). The clinical isolates from the European SENTRY collection were all originally identified on CHROMagar as being C. albicans. However, based on the AFLP patterns shown in Fig. 1, some of these strains were presumably misidentified and belong to different species. When the total collection of isolates previously identified as C. albicans (n = 213) was screened by AFLP analysis, a misidentification rate of 6% was observed. Six strains are now identified as C. dubliniensis, four are identified as C. parapsilosis, one is identified as C. tropicalis, and one is identified as C. guilliermondii (results are partly shown in Fig. 1).
CHROMagar identification of Candida species is based on differences in colony color. It has been shown that the reliability of this method depends on the incubation time and temperature used (2, 24, 35). However, even when optimum conditions are used, the method is not ideal and the differentiation between C. albicans and C. dubliniensis is especially problematic. Kurzai et al. reported that only 81% of their C. dubliniensis isolates showed the dark-green color on CHROMagar, which is considered indicative of C. dubliniensis (17). Furthermore, 15.9% of their C. albicans isolates also showed a dark-green coloration instead of the usual lighter green. Tintelnot et al. (31) reported an even lower number, 57%, of C. dubliniensis isolates that showed the dark-green coloration on CHROMagar, and only 48% of the isolates of Kirkpatrick et al. (15) showing the dark-green coloration turned out to be C. dubliniensis.
Other commercial tests that allow (presumptive) identification of C. albicans as well as non-albicans Candida species usually show high sensitivities and specificities for C. albicans but are less reliable or need further testing for the identification of other, less common species (3, 5, 8, 12). C. dubliniensis-specific PCR assays as well as generic PCR assays in combination with species-specific probes have been developed (6, 7, 16, 19, 25). The advantage of AFLP analysis, however, is that this method is based on the ligation of known sequences (adapters) to restriction fragments, which function as targets for the PCR primers. Therefore, the technique is universally applicable. In the present assay we made use of two subsequent amplifications, but similar results were obtained when only the second amplification was used (unpublished observations). The use of an internal size standard with every sample for normalization purposes greatly enhances the reproducibility between tests. Storing all patterns, including those of the reference strains, in a general, accessible database will provide a screening library for the identification of Candida species.
Two other universally applicable methods for the identification of Candida species have been described: PCR fingerprinting and reference strand-mediated conformational analysis (20, 21). However, whereas PCR fingerprinting uses mini- and microsatellite sequences as targets for the primers and reference strand-mediated conformational analysis is based on 18S rRNA sequences, AFLP patterns are a representation of the whole genome. Our results show very clear differences among medically important Candida species. Therefore, AFLP analysis might prove to be a reliable method for the identification of medically important Candida species, including C. dubliniensis.
Acknowledgments
Annemarie Borst was supported by a grant from bioMérieux.
REFERENCES
- 1.Abi Said, D., E. Anaissie, O. Uzun, I. Raad, H. Pinzcowski, and S. Vartivarian. 1997. The epidemiology of hematogenous candidiasis caused by different Candida species. Clin. Infect. Dis. 24: 1122-1128. [DOI] [PubMed] [Google Scholar]
- 2.Baumgartner, C., A.-M. Freydiere, and Y. Gille. 1996. Direct identification and recognition of yeast species from clinical material by using Albicans ID and CHROMagar Candida plates. J. Clin. Microbiol. 34:454-456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bernal, S., M. E. Martin, M. Chavez, J. Coronilla, and A. Valverde. 1998. Evaluation of the new API Candida system for identification of the most clinically important yeast species. Diagn. Microbiol. Infect. Dis. 32:217-221. [DOI] [PubMed] [Google Scholar]
- 4.Blignaut, E., C. Pujol, S. Lockhart, S. Joly, and D. R. Soll. 2002. Ca3 fingerprinting of Candida albicans isolates from human immunodeficiency virus-positive and healthy individuals reveals a new clade in South Africa. J. Clin. Microbiol. 40:826-836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Campbell, C. K., K. G. Davey, A. D. Holmes, A. Szekely, and D. W. Warnock. 1999. Comparison of the API Candida system with the AUXACOLOR system for identification of common yeast pathogens. J. Clin. Microbiol. 37:821-823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Donnelly, S. M., D. J. Sullivan, D. B. Shanley, and D. C. Coleman. 1999. Phylogenetic analysis and rapid identification of Candida dubliniensis based on analysis of ACT1 intron and exon sequences. Microbiology 145:1871-1882. [DOI] [PubMed]
- 7.Elie, C. M., T. J. Lott, E. Reiss, and C. J. Morrison. 1998. Rapid identification of Candida species with species-specific DNA probes. J. Clin. Microbiol. 36:3260-3265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Espinel-Ingroff, A., L. Stockman, G. Roberts, D. Pincus, J. Pollack, and J. Marler. 1998. Comparison of RapID Yeast Plus System with API 20C system for identification of common, new, and emerging yeast pathogens. J. Clin. Microbiol. 36:883-886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gee, S. F., S. Joly, D. R. Soll, J. F. G. M. Meis, P. E. Verweij, I. Polacheck, D. J. Sullivan, and D. C. Coleman. 2002. Identification of four distinct genotypes of Candida dubliniensis and detection of microevolution in vitro and in vivo. J. Clin. Microbiol. 40:556-574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Giamarellou, H., and A. Antoniadou. 1996. Epidemiology, diagnosis, and therapy of fungal infections in surgery. Infect. Control Hosp. Epidemiol. 17:558-564. [DOI] [PubMed] [Google Scholar]
- 11.Gumbo, T., C. M. Isada, G. Hall, M. T. Karafa, and S. M. Gordon. 1999. Candida glabrata fungemia. Clinical features of 139 patients. Medicine (Baltimore) 78:220-227. [DOI] [PubMed] [Google Scholar]
- 12.Hoppe, J. E., and P. Frey. 1999. Evaluation of six commercial tests and the germ-tube test for presumptive identification of Candida albicans. Eur. J. Clin. Microbiol. Infect. Dis. 18: 188-191. [DOI] [PubMed] [Google Scholar]
- 13.Jones, C. J., K. J. Edwards, S. Castaglione, M. O. Winfield, F. Sala, C. VandeWiel, G. Bredemeijer, B. Vosman, M. Matthes, A. Daly, R. Brettschneider, P. Bettini, M. Buiatti, E. Maestri, A. Malcevschi, N. Marmiroli, R. Aert, G. Volckaert, J. Rueda, R. Linacero, A. Vazquez, and A. Karp. 1997. Reproducibility testing of RAPD, AFLP and SSR markers in plants by a network of European laboratories. Mol. Breed. 3:381-390. [Google Scholar]
- 14.Kao, A. S., M. E. Brandt, W. R. Pruitt, L. A. Conn, B. A. Perkins, D. S. Stephens, W. S. Baughman, A. L. Reingold, G. A. Rothrock, M. A. Pfaller, R. W. Pinner, and R. A. Hajjeh. 1999. The epidemiology of candidemia in two United States cities: results of a population-based active surveillance. Clin. Infect. Dis. 29: 1164-1170. [DOI] [PubMed] [Google Scholar]
- 15.Kirkpatrick, W. R., S. G. Revankar, R. K. Mcatee, J. L. Lopez-Ribot, A. W. Fothergill, D. I. McCarthy, S. E. Sanche, R. A. Cantu, M. G. Rinaldi, and T. F. Patterson. 1998. Detection of Candida dubliniensis in oropharyngeal samples from human immunodeficiency virus-infected patients in North America by primary CHROMagar Candida screening and susceptibility testing of isolates. J. Clin. Microbiol. 36:3007-3012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kurzai, O., W. J. Heinz, D. J. Sullivan, D. C. Coleman, M. Frosch, and F. A. Mühlschlegel. 1999. Rapid PCR test for discriminating between Candida albicans and Candida dubliniensis isolates using primers derived from the pH-regulated PHR1 and PHR2 genes of C. albicans. J. Clin. Microbiol. 37:1587-1590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kurzai, O., H. C. Korting, D. Harmsen, W. Bautsch, M. Molitor, M. Frosch, and F. A. Muhlschlegel. 2000. Molecular and phenotypic identification of the yeast pathogen Candida dubliniensis. J. Mol. Med. 78:521-529. [DOI] [PubMed] [Google Scholar]
- 18.Lott, T. J., and M. M. Effat. 2001. Evidence for a more recently evolved clade within a Candida albicans North American population. Microbiology 147:1687-1692. [DOI] [PubMed] [Google Scholar]
- 19.Martin, C., D. Roberts, M. van der Weide, R. Rossau, G. Jannes, T. Smith, and M. Maher. 2000. Development of a PCR-based line probe assay for identification of fungal pathogens. J. Clin. Microbiol. 38:3735-3742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McIlhatton, B. P., C. Keating, M. D. Curran, M. F. McMullin, J. G. Barr, J. A. Madrigal, and D. Middleton. 2002. Identification of medically important pathogenic fungi by reference strand-mediated conformational analysis (RSCA). J. Med. Microbiol. 51:468-478. [DOI] [PubMed] [Google Scholar]
- 21.Meyer, W., K. Maszewska, and T. C. Sorrell. 2001. PCR fingerprinting: a convenient molecular tool to distinguish between Candida dubliniensis and Candida albicans. Med. Mycol. 39:185-193. [DOI] [PubMed] [Google Scholar]
- 22.Moran, G. P., D. Sanglard, S. M. Donnelly, D. B. Shanley, D. J. Sullivan, and D. C. Coleman. 1998. Identification and expression of multidrug transporters responsible for fluconazole resistance in Candida dubliniensis. Antimicrob. Agents Chemother. 42:1819-1830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Moran, G. P., D. J. Sullivan, M. C. Henman, C. E. McCreary, B. J. Harrington, D. B. Shanley, and D. C. Coleman. 1997. Antifungal drug susceptibilities of oral Candida dubliniensis isolates from human immunodeficiency virus (HIV)-infected and non-HIV-infected subjects and generation of stable fluconazole-resistant derivatives in vitro. Antimicrob. Agents Chemother. 41:617-623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Odds, F. C., and A. Davidson. 2000. “Room temperature” use of CHROMagar Candida. Diagn. Microbiol. Infect. Dis. 38:147-150. [DOI] [PubMed] [Google Scholar]
- 25.Park, S., M. Wong, S. A. E. Marras, E. W. Cross, T. E. Kiehn, V. Chaturvedi, S. Tyagi, and D. S. Perlin. 2000. Rapid identification of Candida dubliniensis using a species-specific molecular beacon. J. Clin. Microbiol. 38:2829-2836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pfaller, M. A., R. N. Jones, G. V. Doern, A. C. Fluit, J. Verhoef, H. S. Sader, S. A. Messer, A. Houston, S. Coffman, R. J. Hollis, et al. 1999. International surveillance of blood stream infections due to Candida species in the European SENTRY Program: species distribution and antifungal susceptibility including the investigational triazole and echinocandin agents. Diagn. Microbiol. Infect. Dis. 35:19-25. [DOI] [PubMed] [Google Scholar]
- 27.Pittet, D., N. Li, and R. P. Wenzel. 1993. Association of secondary and polymicrobial nosocomial bloodstream infections with higher mortality. Eur. J. Clin. Microbiol. Infect. Dis. 12:813-819. [DOI] [PubMed] [Google Scholar]
- 28.Pujol, C., S. Joly, S. R. Lockhart, S. Noel, M. Tibayrenc, and D. R. Soll. 1997. Parity among the randomly amplified polymorphic DNA method, multilocus enzyme electrophoresis, and Southern blot hybridization with the moderately repetitive DNA probe Ca3 for fingerprinting Candida albicans. J. Clin. Microbiol. 35:2348-2358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Savelkoul, P. H. M., H. J. M. Aarts, J. de Haas, L. Dijkshoorn, B. Duim, M. Otsen, J. L. W. Rademaker, L. Schouls, and J. A. Lenstra. 1999. Amplified-fragment length polymorphism analysis: the state of an art. J. Clin. Microbiol. 37:3083-3091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sullivan, D., and D. Coleman. 1998. Candida dubliniensis: characteristics and identification. J. Clin. Microbiol. 36:329-334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tintelnot, K., G. Haase, M. Seibold, F. Bergmann, M. Staemmler, T. Franz, and D. Naumann. 2000. Evaluation of phenotypic markers for selection and identification of Candida dubliniensis. J. Clin. Microbiol. 38: 1599-1608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vos, P., R. Hogers, M. Bleeker, M. Reijans, T. Van de Lee, M. Hornes, A. Frijters, J. Pot, J. Peleman, M. Kuiper, and M. Zabeau. 1995. AFLP: a new technique for DNA fingerprinting. Nucleic Acids Res. 23:4407-4414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wenzel, R. P. 1995. Nosocomial candidemia: risk factors and attributable mortality. Clin. Infect. Dis. 20: 1531-1534. [DOI] [PubMed] [Google Scholar]
- 34.Wey, S. B., M. Mori, M. A. Pfaller, R. F. Woolson, and R. P. Wenzel. 1988. Hospital-acquired candidemia. The attributable mortality and excess length of stay. Arch. Intern. Med. 148:2642-2645. [DOI] [PubMed] [Google Scholar]
- 35.Willinger, B., C. Hillowoth, B. Selitsch, and M. Manafi. 2001. Performance of Candida ID, a new chromogenic medium for presumptive identification of Candida species, in comparison to CHROMagar Candida. J. Clin. Microbiol. 39:3793-3795. [DOI] [PMC free article] [PubMed] [Google Scholar]