Abstract
A peptide-mediated capture PCR for the detection of Mycobacterium avium subsp. paratuberculosis in bulk milk samples was developed and characterized. Capture of the organism was performed using peptide aMptD, which had been shown to bind to the M. avium subsp. paratuberculosis MptD protein (J. Stratmann, B. Strommenger, R. Goethe, K. Dohmann, G. F. Gerlach, K. Stevenson, L. L. Li, Q. Zhang, V. Kapur, and T. J. Bull, Infect. Immun. 72:1265-1274, 2004). Consistent expression of the MptD receptor protein and binding of the aMptD ligand were demonstrated by capturing different Mycobacterium avium subsp. paratuberculosis type I and type II strains and subsequent PCR analysis using ISMav2-based primers. The analytical sensitivity of the method was determined to be 5 × 102 CFU ml−1 for artificially contaminated milk. The specificity of aMptD binding was confirmed by culture and competitive capture assays, showing selective enrichment of M. avium subsp. paratuberculosis (at a concentration of 5 × 102 CFU ml−1) from samples containing 100- and 1,000-fold excesses of other mycobacterial species, including M. avium subsp. avium and M. avium subsp. hominissuis. The aMptD-mediated capture of M. avium subsp. paratuberculosis using paramagnetic beads, followed by culture, demonstrated the ability of this approach to capture viable target cells present in artificially contaminated milk. Surface plasmon resonance experiments revealed that the aMptD peptide is a high-affinity ligand with a calculated association rate constant of 9.28 × 103 and an association constant of 1.33 × 109. The potential use of the method on untreated raw milk in the field was investigated by testing 423 bulk milk samples obtained from different dairy farms in Germany, 23 of which tested positive. Taken together, the results imply that the peptide-mediated capture PCR might present a suitable test for paratuberculosis screening of dairy herds, as it has an analytical sensitivity sufficient for detection of M. avium subsp. paratuberculosis in bulk milk samples under field conditions, relies on a defined and validated ligand-receptor interaction, and is adaptable to routine diagnostic laboratory automation.
Paratuberculosis (Johne's disease) is a chronic and incurable granulomatous enteritis of ruminants caused by Mycobacterium avium subsp. paratuberculosis (29). The disease occurs worldwide with increasing frequency (32) and has a considerable economic impact on the livestock industry (21). Calves are mostly infected in early life, with high shedding and clinical disease commonly occurring at 2 to 5 years of age (37). Previous attempts to eradicate the disease from infected herds have frequently failed, despite a high degree of organizational and financial efforts (3, 26). This is most likely due to the ubiquitous presence of the pathogen in the environments of infected herds (39) and its very great tenacity (7, 20, 52).
An economically feasible alternative to eradication would be a control program aiming at the early identification and removal of high shedders, thereby reducing environmental contamination and infectious pressure on the herd. High shedders are more likely to secrete M. avium subsp. paratuberculosis in milk (50), and in addition, in herds with high shedders the pathogen is more likely to enter the milk by fecal contamination (8). Therefore, bulk milk might be a suitable diagnostic substrate for such an approach. Since the general infrastructure for testing of bulk milk from farms (i.e., untreated raw milk) is established (25), regular testing of this milk for the presence of M. avium subsp. paratuberculosis could allow early detection of herds with high shedders. The most convenient and amenable methods for such detection of M. avium subsp. paratuberculosis DNA in milk are enrichment via immunomagnetic separation (17, 28, 38) and peptide-mediated capture (47) followed by PCR, as these methods can be adapted to high-throughput testing using standard laboratory automation.
In the study presented here, we followed this approach, using the phage display-derived peptide aMptD (46) for the capture of M. avium subsp. paratuberculosis in milk samples. The aMptD peptide was shown to bind to the surface-exposed MptD protein of M. avium subsp. paratuberculosis (ORF 3733C) (31), which is part of an M. avium subsp. paratuberculosis-specific pathogenicity island (46), and therefore this method, in contrast to the previously described peptide-mediated capture assay (47), is based on a defined receptor-ligand interaction. Furthermore, we thoroughly elucidated the strain and species cross-specificity of peptide aMptD for M. avium subsp. paratuberculosis by performing competitive capture assays, and we determined the kinetics and affinity of the receptor-ligand interaction by surface plasmon resonance (SPR) (27) in BIAcore biosensor experiments. Finally, in order to investigate the potential use of the method in the field, we applied the peptide aMptD-mediated capture assay with subsequent PCR analysis (incorporating an internal amplification control [IAC]) to 423 bulk milk samples obtained from German dairy farms.
MATERIALS AND METHODS
Bacterial strains, plasmids, primers, and growth conditions.
The bacterial strains, plasmids, primers, and peptides used in this study are listed in Tables 1 and 2. Mycobacteria were grown on BBL Herrold's egg yolk agar slants (Becton Dickinson, Sparks, Md.) or on Middlebrook 7H10 agar (Difco Laboratories, Detroit, Mich.) supplemented with 10% oleic acid-albumin-dextrose-catalase enrichment (100 ml contained sodium chloride [145 mM], bovine serum albumin [fraction V; 5 g], dextrose [1.1 M], catalase [3 mg], and oleic acid [60 μl]), glycerol (0.2%), and mycobactin J (2 μg liter−1; Synbiotics, Lyon, France). For further use in the capture assays, mycobacteria were harvested from Middlebrook 7H10 agar by careful removal from the agar and resuspended in phosphate-buffered saline (NaCl [150 mM], KH2PO4 [1.5 mM], Na2HPO4 [9 mM], KCl [2.5 mM], pH 7.2). Homogenization of the suspension was done by vortexing it for 5 minutes with glass beads (30 beads with a 3-mm diameter per 5 ml of bacterial suspension in a polypropylene tube). Plasmid-carrying Escherichia coli strains were grown in Luria-Bertani medium supplemented with ampicillin (100 μg ml−1).
TABLE 1.
Strains used in this study
| Species or strain | Description | Source |
|---|---|---|
| Mycobacterium species | ||
| M. avium subsp. paratuberculosis | Strain 6783 (DSM 44135) | Laboratory reference strain (clinical isolate) |
| M. avium subsp. avium | DSM 44156/ATCC 25291T | German Collection of Microorganisms and Cell Cultures (DSMZ) |
| Two M. avium subsp. hominissuis clinical isolates | Clinical isolates | I. Moser, Friedrich-Loeffler-Institut, Jena, Germany |
| Six M. avium subsp. paratuberculosis field isolates | EU reference panel no. 25 and 30 (type I strains) and no. 4, 6, 40, and 49 (type II strains) | K. Stevenson, Moredun Research Institute, Penicuik, Scotland |
| M. fortuitum | DSM 46621 | I. Moser, Friedrich-Loeffler-Institut, Jena, Germany |
| M. diernhoferi | DSM 43524 | I. Moser, Friedrich-Loeffler-Institut, Jena, Germany |
| M. vaccae | Field isolate | I. Moser, Friedrich-Loeffler-Institut, Jena, Germany |
| M. flavescens | DSM 43991 | I. Moser, Friedrich-Loeffler-Institut, Jena, Germany |
| M. gastri | Field isolate | I. Moser, Friedrich-Loeffler-Institut, Jena, Germany |
| M. gordonae | DSM 43212 | I. Moser, Friedrich-Loeffler-Institut, Jena, Germany |
| M. malmoense | DSM 44163 | I. Moser, Friedrich-Loeffler-Institut, Jena, Germany |
| E. coli strain | ||
| TOP10 F′ | F′ mcrA Δ(mrr-hsdRMS-mcrBC) φ80dlacZΔM15 ΔlacX74 recA1 deoR araD139 Δ(ara leu)7697 galU galK rpsL StrrendA1 nupG | TOPO TA cloning kit (Invitrogen, Groningen, The Netherlands) |
TABLE 2.
Plasmids, primers, and peptides used in this study
| Plasmid, primer, or peptide | Description or sequence (5′-3′) | Reference or source |
|---|---|---|
| Plasmids | ||
| pCR 2.1 TOPO | Topoisomerase I “enhanced” E. coli cloning vector carrying ampicillin and kanamycin resistance determinants as well as the lacZ gene for blue-white selection | TOPO TA cloning kit (Invitrogen, Groningen, The Netherlands) |
| pMAPC | pCR 2.1 TOPO containing a 402-bp fragment of the M. avium subsp. paratuberculosis mptF gene flanked by specific 20-mers | This work |
| Primers | ||
| ISMav1 | GTA TCA GGC CGT GAT GGC GG | 47 |
| ISMav2 | CCG CAC CAG CGC TCG ATA CA | 47 |
| oMapC long1 | ACA CTT GCG ATC TGG GCC TCT TGA GGT AAG CAA GTT CGC GCC | This work |
| oMapC long2 | ATC GCG CTA CCC ATT CGG TGA TCA TGG CTG AGG TAG TCG GC | This work |
| oMapC1 | ACA CTT GCG ATC TGG GCC TCT | This work |
| oMapC2 | ATC GCG CTA CCC ATT CGG TGA | This work |
| oABC5 | TAG CTC GAC GAC CCA GTC CA (mptD-derived forward primer situated at positions 4861 to 4881 of the mpt operon [GenBank accession no. AF419325]) | This work |
| oABC6 | TCA GCC CGA ATA CCA CCG TG (mptD-derived reverse primer situated at positions 5395 to 5415 of the mpt operon [GenBank accession no. AF419325]) | This work |
| MK5 | TTC TTG AAG GGT GTT CGG GGC C (IS900-derived forward primer) | 10 |
| MK6 | GCG ATG ATC GCA GCG TCT TTG G (IS900-derived reverse primer) | 10 |
| MK7 | GTC TGG GAT TGG ATG TCC TG (IS901-derived forward primer) | 30 |
| MK8 | CAC CAC GTG GTT AGC AAT CC (IS901-derived reverse primer) | 30 |
| P1-IS1245 | GCC GCC GAA ACG ATC TAC (IS1245-derived forward primer) | 18 |
| P2-IS1245 | AGG TGG CGT CGA GGA AGA (IS1245-derived reverse primer) | 18 |
| P264 | TGC ACA CAG GCC ACA AGG GA (M. avium subsp. paratuberculosis 16S RNA gene-derived reverse primer situated at positions 1015 to 1034) | 6 |
| P283 | GAG TTT GAT CCT GGC TCA GGA (M. avium subsp. paratuberculosis 16S RNA gene-derived forward primer situated at positions 11 to 31) | E. C. Boettger, personal communication |
| Peptides | ||
| aMptD | GKNHHHQHHRPQ | 46 |
| aMpR | HSQPKQVKKASR | 46 |
Determination of mycobacterial cell counts.
The optical densities at 600 nm (OD600) of the mycobacterial suspensions used for the capture assays were measured, viability was confirmed using the BacLight system (Invitrogen, Groningen, The Netherlands), and the numbers of CFU were determined by plating serial 10-fold dilutions on Middlebrook 7H10 agar supplemented with 10% oleic acid-albumin-dextrose-catalase enrichment, 0.2% glycerol, and mycobactin J (2 μg liter−1) after an incubation period of 6 weeks at 37°C.
Manipulation and analysis of DNA.
Agarose gel electrophoresis, plasmid preparation, PCR, DNA cloning, and transformation of E. coli were done by standard procedures (41). DNA-modifying enzymes were purchased from New England Biolabs (Frankfurt, Germany) and used according to the manufacturer's instructions. Taq polymerase and primers were purchased from Invitrogen (Groningen, The Netherlands). Sequencing reactions were done by SeqLab GmbH (Goettingen, Germany), and sequencing data analyses were performed with the Husar 5.0 program (DKFZ, Heidelberg, Germany).
Construction of internal control for capture PCR.
An internal amplification control designed to be amplified under the same PCR conditions as those described for the ISMav2-derived primer set was constructed. In the first step, a 402-bp fragment of the M. avium subsp. paratuberculosis mptF gene (46) was amplified using two 40-mer primers (oMapC long1 and -2), each consisting of an mptF-specific 20-mer oligonucleotide and a 5′ random 20-mer oligonucleotide, which do not result in an amplification product when used with Mycobacterium avium subsp. paratuberculosis DNA. The complete fragment was then cloned into the pCR2.1-TOPO vector (Invitrogen, Groningen, The Netherlands) and transformed into E. coli Top10F′ cells according to the manufacturer's instructions, and mutants were selected using blue-white selection. White colonies were screened for the presence of an insert by PCR using the 20-mer oligonucleotide primers oMapC1 and -2 (Table 1). Plasmid DNA of a PCR-positive clone was extracted with a NucleoBond AX kit (Machery Nagel, Dueren, Germany), confirmed by nucleotide sequence analyses, and designated pMAPC. The optimal concentrations of the internal control plasmid pMAPC and primers oMapC1 and -2 were titrated in a series of PCRs containing defined serial dilutions of M. avium subsp. paratuberculosis DNA. This resulted in a PCR master mix containing 5 pmol each of oMapC1 and -2, 25 pmol each of ISMav1 and ISMav2, and 0.1 pg pMAPC per 50-μl PCR mix.
Peptide aMptD-mediated capture PCR.
The aMptD peptide (GKNHHHQHHRPQ) was synthesized (Fa. Affina Immuntech, Berlin, Germany) and coupled directly (i.e., without a biotin-streptavidin bridge) to paramagnetic beads by the carbodiimide method (Fa. Chemicell, Berlin, Germany). Carbodiimides react with the terminal carboxylate groups from the magnetic beads to form highly reactive O-acylisourea derivatives, which then form a covalent bond with the free amino group of the lysine of peptide aMptD. Peptide-mediated capture PCR and spiking of pasteurized milk were performed essentially as described previously (47) but with some modifications allowing automated processing. Briefly, bulk milk samples (1 ml) were distributed in deep-well plates (Abgene, Epsom, United Kingdom); 5 μg of peptide aMptD-coupled paramagnetic beads (suspended in 10 μl of distilled water) was added to each well, and each deep-well plate was incubated overnight at 4°C with slight agitation. Beads were sedimented using a MagnaBot96 magnetic separation device (Promega, Madison, WI), and the milk was removed; the beads were resuspended in 200 μl of 0.1× Tris-buffered saline buffer (50 mM Tris-HCl [pH 7.5], 150 mM NaCl) containing 0.05% Tween 20 and transferred to U-bottomed microtiter plates. In these plates, beads were magnetically sedimented and washed 10 times with 0.1× Tris-buffered saline-Tween (200 μl) using a Nunc-Immuno washer (Nunc, Roskilde, Denmark). After the final wash, beads were resuspended in 50 μl 0.1× TE buffer (1 ×TE buffer is 10 mM Tris-HCl [pH 8.0] and 1 mM EDTA) and boiled in a microwave oven for 15 min at 180 W. After the beads were sedimented again, the supernatant was transferred to a new microtiter plate, and 5 μl of the supernatant was used as a template in a PCR using primers ISMav1 and -2. The specificity of the PCR was confirmed by restriction enzyme digestion or by sequencing of the products. The bulk milk samples tested in this study were kindly provided by several dairy plants in eastern and northern Germany.
Preparation of mycobacterial membranes.
M. avium subsp. paratuberculosis and M. avium subsp. avium strains were grown on Middlebrook 7H10 agar; bacteria were harvested by careful removal from the agar, and approximately 100 mg (wet weight) was resuspended in 20 ml Tris-HCl (30 mM [pH 8.0]). Bacteria were pelleted by centrifugation (4,000 × g for 10 min) and resuspended in 2 ml Tris-HCl (30 mM [pH 8.0]) containing 20% saccharose. Bacteria were disrupted mechanically with zirconium beads (200 μg ml−1; Carl Roth GmbH, Karlsruhe, Germany) in a Fast Prep instrument (QBiogene, Heidelberg, Germany) for 90 seconds at the highest intensity. Subsequently, suspensions were sonicated (3 times for 30 s each at output 2 or 3, using a sonicator minitip [Sonic Cell Disruptor; Branson Power Co., Danbury, CT]). Cell debris and zirconium beads were sedimented via centrifugation (11,000 × g for 10 min) and discarded. The supernatant was transferred to an ultracentrifugation tube, the volume was adjusted to 7.5 ml with Tris-HCl (30 mM [pH 8]) containing 20% sucrose, and the mixture was centrifuged in a type 65 rotor (Sorvall Ultra Pro; DuPont Instruments, Bad Homburg, Germany) at 177,700 × g for 2 h at 4°C. The pellet was dissolved in 500 μl TE buffer and centrifuged at 13,000 rpm in an Eppendorf centrifuge at room temperature to remove aggregates. The supernatant remaining after this centrifugation step contained the mycobacterial membranes. Membrane preparations were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis as described earlier (15). The protein concentration was determined by using a MicroBCA assay (Uptima Interchim, Montlucon Cedex, France).
Surface plasmon resonance analyses.
Surface plasmon resonance analyses were done using a BIAcore 3000 instrument (Biacore International SA, Neuchatel, Switzerland), and data were analyzed using BIAevaluation software, version 3.0. The running buffer used for all experiments was HEPES-buffered saline containing HEPES (10 mM [pH 7.4]), NaCl (150 mM), EDTA (3 mM), and surfactant P20 (0.005%; Biacore International SA, Neuchatel, Switzerland). Research-grade CM5 chips (Biacore International SA) were activated by injecting a mix containing equal parts of N-ethyl-N-dimethylaminopropyl-carbodiimide and N-hydroxysuccinimide according to the manufacturer's instructions. Mycobacterium avium subsp. paratuberculosis and M. avium subsp. avium membranes (100 μg ml−1) were immobilized on the chips in sodium acetate buffer (10 mM [pH 4.0]) at a flow rate of 10 μl min−1, followed by an injection of ethanolamine (1 M [pH 8.5]) to block free sites on the dextran matrix. This treatment resulted in 50 (M. avium subsp. avium) to 75 (M. avium subsp. paratuberculosis) resonance units (RU) for the immobilized membranes. The chip flow cells were set up as follows: channels 1 and 3 were used as blanks (reference flow cells), M. avium subsp. paratuberculosis membranes were bound to channel 2, and M. avium subsp. avium membranes were bound to channel 4. For determinations of binding affinity, twofold serial dilutions of the aMptD peptide and the negative control peptide aMpR (HSQPKQVKKASR) resulting in concentrations of 3.12 to 25 μg ml−1 were injected at a flow rate of 10 μl min−1 for 5 min. The sensor chip was regenerated between cycles with a 1-minute injection of glycine-HCl (10 mM [pH 2.5]) at the same flow rate. The binding of peptides was monitored and presented in a sensogram (a plot of RU versus time); a 0.1° shift in the SPR angle, corresponding to 1,000 RU, corresponds to a change in the surface concentration of 1 ng mm−2 (13). The RU responses for interactions of the aMptD peptide with M. avium subsp. paratuberculosis and M. avium subsp. avium membranes were calculated by subtracting the respective RU values obtained at the same peptide concentration upon injection over the reference flow cells (blank). The RU responses for the interactions of the control peptide aMpR with the mycobacterial membranes were calculated using the same setup as that for peptide aMptD. To calculate the association rate and dissociation rate constants (kas and kdis) and the association constant (KA = kdis/kas) of the aMptD peptide, the resulting sensograms were plotted, and kinetic data were calculated using the Langmuir 1:1 model included in the BIAeval software.
RESULTS
Cross-strain reactivity of peptide aMptD for different M. avium subsp. paratuberculosis strains.
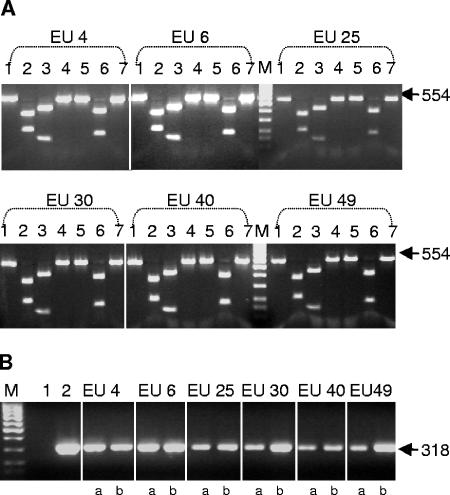
Peptide aMptD has been shown to bind to the surface-exposed protein MptD of Mycobacterium avium subsp. paratuberculosis strain 6783. In order to analyze whether the aMptD peptide would have the potential to bind the MptD proteins of different M. avium subsp. paratuberculosis isolates, the mptD genes of four type II and two type I strains were PCR amplified, and the products were analyzed by restriction enzyme digestion. Digests of the PCR products with AvaI, BsrBI, HinfI, NarI, NgoMIV, and MseI resulted in identical restriction endonuclease profiles for all strains, thereby indicating that the mptD genes in M. avium subsp. paratuberculosis type I and II strains are identical (Fig. 1A). To confirm the binding of the aMptD peptide to different M. avium subsp. paratuberculosis strains, the peptide was coupled to paramagnetic beads and incubated with milk spiked with 5 × 102 and 5 × 103 CFU ml−1 of these strains. Subsequent PCRs with primers ISMav1 and 2 demonstrated the capture of the different strains with peptide aMptD (Fig. 1B). This finding showed that the MptD protein is highly conserved among different M. avium subsp. paratuberculosis isolates and thereby confirmed that the aMptD peptide is suitable for use as a ligand in a diagnostic test.
FIG. 1.
Restriction enzyme analysis and peptide aMptD-mediated capture PCR with different M. avium subsp. paratuberculosis strains. (A) Restriction enzyme digestion of the PCR-amplified mptD gene. Lanes 1, undigested PCR product; lanes 2, digest with AvaI; lanes 3, digest with BsrBI; lanes 4, digest with HinfI; lanes 5, digest with NarI; lanes 6, digest with NgoMIV; lanes 7, digest with MseI; lanes M, 100-bp DNA marker. The arrows to the right indicate the expected position of the undigested PCR product (in base pairs). (B) Peptide aMptD-mediated capture PCR with different M. avium subsp. paratuberculosis strains from artificially contaminated milk, using ISMav2-derived primers in spiked milk. Lane 1, negative control; lane 2, positive control; lane M, 100-bp DNA marker. Mycobacteria were spiked at different concentrations (a, 5 × 103 ml−1; b, 5 × 102 ml−1). The arrow to the right indicates the expected position of the PCR product (in base pairs).
Specificity of peptide aMptD for M. avium subsp. paratuberculosis.
Milk was spiked with M. avium subsp. paratuberculosis 6783 at concentrations of 5 × 105 down to 5 × 100 CFU ml−1 and incubated with aMptD-coated paramagnetic beads or uncoated beads. After overnight incubation, the beads were sedimented, washed, and spread on Herrold's egg yolk agar slants containing mycobactin J. After 6 weeks of incubation, colonies were visible on all slants inoculated with the aMptD-coated beads, whereas no colonies were visible on the slants inoculated with uncoated beads. The colonies were confirmed to be M. avium subsp. paratuberculosis by PCR using ISMav2- and IS900-derived primers. These experiments demonstrated that capture and subsequent cultivation of viable M. avium subsp. paratuberculosis isolates are possible using aMptD-coated beads.
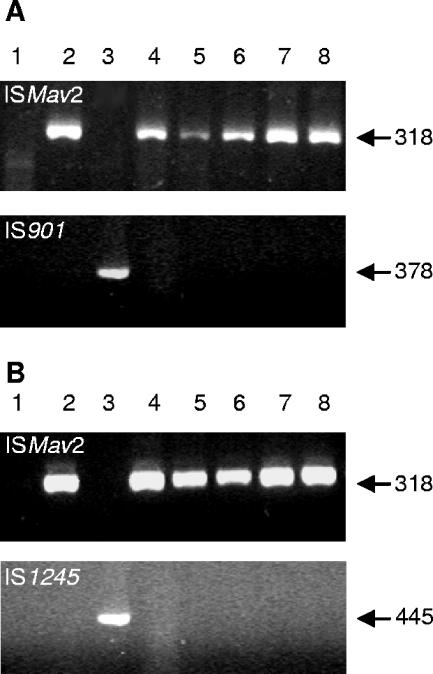
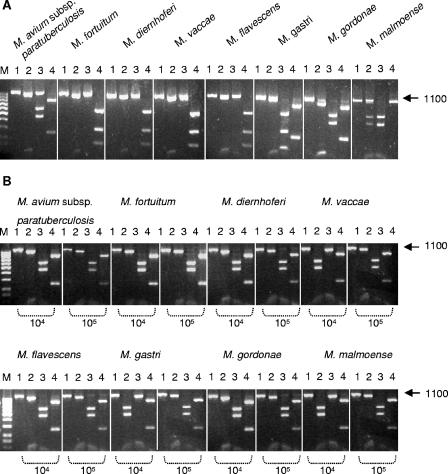
To further confirm the M. avium subsp. paratuberculosis specificity of the peptide, a competitive peptide-mediated capture assay was performed with milk spiked simultaneously with 5 × 103 CFU ml−1 M. avium subsp. paratuberculosis and 5 × 105 down to 5 × 100 CFU ml−1 M. avium subsp. avium or M. avium subsp. hominissuis. After overnight capture, only M. avium subsp. paratuberculosis was detected by PCR analysis using ISMav2-derived primers; with IS901-derived primers, no M. avium subsp. avium could be detected (Fig. 2A). Similarly, with IS1245-derived primers, no M. avium subsp. hominissuis could be detected (Fig. 2B). In addition, competitive capture assays with seven other mycobacterial species (Mycobacterium fortuitum, Mycobacterium diernhoferi, Mycobacterium vaccae, Mycobacterium flavescens, Mycobacterium gastri, Mycobacterium gordonae, and Mycobacterium malmoense) were performed to demonstrate the specificity of the aMptD peptide for M. avium subsp. paratuberculosis. In order to distinguish whether M. avium subsp. paratuberculosis or the competing mycobacteria had been captured, the subsequent PCR assay was based on amplification of the 16S RNA gene followed by digestion with the restriction endonucleases AvaI, NciI, and Tsp451 (Fig. 3A; Table 3). For the competitive capture assay, milk was spiked simultaneously with 5 × 102 CFU ml−1 M. avium subsp. paratuberculosis and a 100- or 1,000-fold excess of one of the seven different mycobacterial species. Overnight capture followed by PCR and restriction endonuclease analyses clearly demonstrated that solely M. avium subsp. paratuberculosis had been captured (Fig. 3B) and thereby confirmed the species specificity of the aMptD peptide.
FIG. 2.
Competitive capture PCR amplification from artificially contaminated milk. Milk was artificially contaminated with a mixture of M. avium subsp. avium and M. avium subsp. paratuberculosis (A) or M. avium subsp. hominissuis and M. avium subsp. paratuberculosis (B). Lanes 1, negative control; lanes 2, M. avium subsp. paratuberculosis DNA; lanes 2, M. avium subsp. avium (A) or M. avium subsp. hominissuis DNA (B); lanes 4 to 8, capture from milk spiked with 105 to 101 bacteria ml−1 of M. avium subsp. avium (A) or M. avium subsp. hominissuis (B), with each sample containing 103 bacteria ml−1 of M. avium subsp. paratuberculosis. The arrows to the right indicate the expected positions of the PCR products obtained with ISMav2-derived primers (top), IS901-derived primers (bottom in panel A), or IS1245-derived primers (bottom in panel B).
FIG. 3.
Restriction enzyme analysis of PCR-amplified 16S RNA genes of different mycobacterial strains and competitive capture PCR amplification from milk artificially contaminated with a mixture of M. avium subsp. paratuberculosis and seven different mycobacterial species. (A) Restriction enzyme digestion of the PCR-amplified 16S RNA genes of the mycobacterial species indicated at the top. Lanes 1, undigested PCR product; lanes 2, digest with AvaI; lanes 3, digest with NciI; lanes 4, digest with Tsp45I; lane M, 100-bp DNA marker. The arrow on the right indicates the expected position of the PCR product (in base pairs). (B) Competitive capture PCR amplification from milk artificially contaminated with a mixture of M. avium subsp. paratuberculosis and seven different mycobacterial species, followed by restriction enzyme digestion. Lanes 1, undigested PCR product; lanes 2, digest with AvaI; lanes 3, digest with NciI; lanes 4, digest with Tsp45I. The number of bacteria ml−1 (104 or 105) of each mycobacterial species in spiked milk is indicated, with each sample containing 102 bacteria ml−1 of M. avium subsp. paratuberculosis. Lane M, 100-bp DNA marker. The arrow to the right indicates the expected position of the PCR product (in base pairs).
TABLE 3.
Expected fragment sizes after restriction enzyme digestion of PCR products from 16S RNA genes of various mycobacteria
| Species | Fragment size(s) (bp)
|
||
|---|---|---|---|
| NciI | Tsp45I | AvaI | |
| M. avium subsp. paratuberculosis | 648, 452 | 846, 254 | 1,010, 90 |
| M. fortuitum | 1,100 | 156, 267, 677 | 1,010, 90 |
| M. diernhoferi | 1,100 | 156, 267, 677 | 1,010, 90 |
| M. vaccae | 1,100 | 153, 266, 681 | 1,010, 90 |
| M. flavescens | 980, 120 | 156, 267, 677 | 1,010, 90 |
| M. gastri | 676, 313, 111 | 874, 226 | 1,010, 90 |
| M. malmoense | 656, 444 | 854, 246 | 928, 90, 82 |
| M. gordonae | 656, 444 | 1,100 | 1,010, 90 |
Investigation of the binding affinity of peptide aMptD for M. avium subsp. paratuberculosis membranes.
In order to investigate the affinity of the aMptD peptide for isolated M. avium subsp. paratuberculosis membranes, SPR analysis using the BIAcore system was performed. SPR allows the semiquantitative analysis of interactions between ligand molecules in solution and receptor molecules attached to a sensor chip surface, providing a continuous readout of the ligand-receptor interaction over time. Sensograms of the membrane-peptide interactions were monitored as shown in Fig. 4. Comparison of the different sensograms obtained indicated a distinct increase in RU (up to 340 RU) when aMptD was passed over M. avium subsp. paratuberculosis membranes, whereas the increase in RU was considerably lower (up to a maximum of only 50 RU) when aMptD was injected over M. avium subsp. avium membranes. The increase in RU observed when aMptD was injected over M. avium subsp. paratuberculosis membranes was proportional to the concentration of peptide injected. Injection of the control peptide over the immobilized membranes did not result in a measurable increase in RU. Sensograms were plotted, and the kinetic data were calculated using the Langmuir 1:1 model, resulting in affinity and dissociation constants for the aMptD peptide. The calculated constants obtained for aMptD interaction with M. avium subsp. paratuberculosis membranes revealed an association rate constant (kas) of 9.28 × 103 and an association constant (KA) of 1.33 × 109, and for aMptD interaction with M. avium subsp. avium, an association rate constant (kas) of 1.16 × 103 and an association constant (KA) of 6.38 × 106 were calculated (Table 4). The high affinity of aMptD suggests that it will be useful as a capture ligand in diagnostic assays for the detection of M. avium subsp. paratuberculosis.
FIG. 4.
Plotted sensograms obtained using the BIAcore system. Peptides aMptD and aMpR were injected at increasing concentrations (3.125 μg ml−1, 6.25 μg ml−1, 12.5 μg ml−1, and 25 μg ml−1) over immobilized membranes of M. avium subsp. paratuberculosis and M. avium subsp. avium.
TABLE 4.
Binding constants for peptide aMptD, determined by numerical integration using the Langmuir 1:1 modela
| Species | aMptD binding constant
|
|||
|---|---|---|---|---|
| kas (1/M · s) | kdis (1/s) | KA (1/M) | KD (M) | |
| M. avium subsp. paratuberculosis | 9.28 × 103 | 6.97 × 10−6 | 1.33 × 109 | 7.51 × 10−10 |
| M. avium subsp. avium | 1.16 × 103 | 1.81 × 10−4 | 6.38 × 106 | 1.57 × 10−7 |
kas, association rate constant; kdis, dissociation rate constant; KA, association constant (kdis/kas); KD, dissociation constant (kas/kdis).
Analytical sensitivity of peptide aMptD-mediated capture PCR and its use for the detection of Mycobacterium avium subsp. paratuberculosis in bulk milk samples.
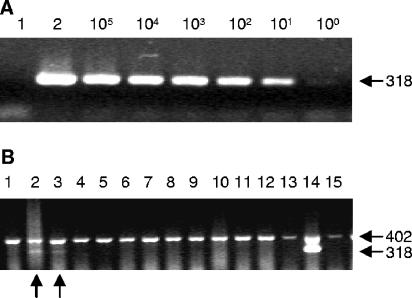
In order to investigate the analytical sensitivity of the method, pasteurized milk was spiked with serial 10-fold dilutions of M. avium subsp. paratuberculosis starting at a 1-to-100 dilution of a suspension with an OD600 of 0.1; viability staining and plating on Middlebrook agar showed that an OD600 of 0.1 translates to a concentration of 5 × 107 CFU ml−1. After peptide-mediated capture from 1-ml volumes, M. avium subsp. paratuberculosis was detected in a PCR with primers ISMav1 and -2 at a concentration of 5 × 102 CFU ml−1 or below (Fig. 5A). The stability and reproducibility of the entire approach were confirmed by repeating the experiment with 10 different bead preparations, using different Mycobacterium avium subsp. paratuberculosis preparations each time; a minimal analytical sensitivity of 5 × 102 CFU ml−1 was consistently achieved, and it proved to be unaffected by the introduction of an IAC designed to avoid false-negative results.
FIG. 5.
Peptide aMptD-mediated capture PCR. (A) Peptide-mediated capture PCR with artificially contaminated milk, using ISMav2-derived primers. Lane 1, negative control; lane 2, positive amplification control with M. avium subsp. paratuberculosis DNA as the template. The number (105 to 100) of M. avium subsp. paratuberculosis bacteria ml−1 is indicated. The arrow on the right indicates the expected position of the PCR product (in base pairs). (B) Peptide-mediated capture PCR amplification from farm-derived bulk milk samples, using ISMav2-specific primers and incorporated IAC. Lanes 1 to 13, peptide-mediated capture PCR amplification from farm-derived bulk milk samples; lane 14, positive control (milk spiked with 102 bacteria of M. avium subsp. paratuberculosis ml−1); lane 15, negative control. The arrows on the right indicate the expected positions of the PCR products (in base pairs); the arrows on the bottom indicate the positions of positive bulk milk samples.
In order to investigate the applicability of the method to untreated raw milk, 423 bulk milk samples obtained from different dairy herds throughout Germany were tested. Twenty-three of the samples (5.5%) were positive by PCR, and 29 (6.8%) of the PCRs failed (i.e., no PCR fragment was obtained from the IAC). An example of a diagnostic PCR with the IAC is shown in Fig. 5B. In order to verify the identity of the ISMav2 PCR-derived DNA fragments, five amplicons obtained from positive bulk milk samples were confirmed by nucleotide sequence analysis, showing that the peptide aMptD-mediated capture PCR is applicable to untreated milk under routine conditions in a diagnostic laboratory.
DISCUSSION
In this report, we present the development and characterization of a peptide-mediated capture PCR for the detection of M. avium subsp. paratuberculosis in bulk milk samples from dairy herds. A significant improvement compared to the method we described previously (47) was achieved by using the peptide aMptD as the ligand (46). The aMptD peptide could be coupled directly to paramagnetic beads without a biotin-streptavidin linker, and the capture of the organism is now based on a defined receptor-ligand interaction. We confirmed the analytical sensitivity of the approach, extensively validated its specificity, and incorporated an IAC into the subsequent PCR analysis to prevent the occurrence of false-negative results. Finally, we tested the optimized protocol under field conditions by applying it to 423 bulk milk samples.
Currently, mandatory paratuberculosis control programs for dairy cattle are rare (2); this is likely due to a lack of reliable and reasonably priced diagnostic methods such as those which have been developed for other infectious diseases, using bulk milk as the diagnostic substrate (5). The milk-based programs in place rely on an enzyme-linked immunosorbent assay (ELISA) of individual milk samples (22, 36). However, despite specificities of up to 99% (24, 40), false-positive reactions are bound to occur, particularly in low-prevalence herds, leading to poor acceptance of detect-and-cull strategies by farmers. In addition, not all shedders can be detected based on an antibody response (44). The situation could be improved considerably if a bulk milk-based and highly specific diagnostic test relying on direct detection of the pathogen itself were available. Various studies have shown that this goal can be achieved in principle, effectively based on immunomagnetic (16, 17, 28) or peptide-mediated (47) enrichment of the pathogen from milk followed by subsequent PCR analysis.
The aMptD peptide was shown previously to bind to the surface-exposed MptD protein of M. avium subsp. paratuberculosis strain 6783 (46). However, it was shown that the expression profiles of proteins can vary between different M. avium subsp. paratuberculosis and M. avium subsp. avium strains (4, 51). Therefore, in order to investigate the suitability of the aMptD peptide as a ligand for diagnostic purposes, the presence of the mptD gene in six different M. avium subsp. paratuberculosis type I and II strains was tested by PCR analysis. Subsequent restriction endonuclease digests confirmed that the gene was highly conserved among the different isolates tested. The consistent expression of the MptD protein and the cross-strain reactivity of peptide aMptD were then tested by capturing two M. avium subsp. paratuberculosis type I and four type II strains. It was shown that, independent of the strain, 5 × 102 ml−1 organisms of M. avium subsp. paratuberculosis could be reliably detected using the aMptD-based capture PCR.
Besides cross-strain reactivity, species specificity is a major concern for diagnostic reagents. Previous studies had merely demonstrated the absence of the mptD gene from a closely related M. avium subsp. avium reference strain, and it was shown that the 38-kb pathogenicity island which contains the mptD gene is M. avium subsp. paratuberculosis specific (42). To additionally ensure that no cross-reactive receptors are present in M. avium subsp. avium, M. avium subsp. hominissuis, or other mycobacteria, competitive capture assays were performed. The finding that 5 × 102 M. avium subsp. paratuberculosis cells were consistently detected whereas no other mycobacteria could be captured, even in the presence of a 2- to 3-log excess, clearly demonstrated the species specificity of the MptD protein receptor.
Since peptides are not commonly used in diagnostic tests to date, the affinity of the receptor-ligand interaction had to be ensured. For this purpose, SPR presents the most adequate technique, as it was shown that SPR is a suitable tool for examining receptor-ligand interactions (1), estimating antibody specificities (14), and calculating membrane-binding affinities of peptides (19). Based on these findings, we chose SPR to quantify the aMptD-receptor interactions. The difference in the association constants revealed an explicitly higher affinity of peptide aMptD for M. avium subsp. paratuberculosis than for M. avium subsp. avium. The values of the association rate constant kas (9.28 × 103) and the association constant KA (1.33 × 109) are comparable to those described for high-affinity antibodies or peptides (11, 35). These results demonstrate that the MptD peptide binds to M. avium subsp. paratuberculosis with the high affinity required for diagnostic reagents.
After showing that the aMptD peptide is highly specific, we tested its diagnostic applicability as a ligand for the capture of M. avium subsp. paratuberculosis from milk samples. First, using artificially contaminated milk, we found that captured bacteria remained viable. Furthermore, initial tests of the capture PCR with spiked milk confirmed an analytical sensitivity of 5 × 102 CFU ml−1, which is comparable to the immunomagnetic separation results of previously reported studies (17, 34). The PCR used was based on primers directed against the specific ISMav2 insertion element (49); this element has been shown to be specific (43), whereas for PCRs based on the IS900 element (33, 45, 48), false-positive reactions have been reported (9, 12). An IAC was included in order to prevent the occurrence of false-negative results, and thereby the significance of the PCR results obtained was improved (23). Since the capture PCR should be utilized for the screening of large numbers of bulk milk samples, it needed to be adapted to routine laboratory automation. In contrast to the case for previously described immunomagnetic separation protocols (16), we could overcome the problem of remaining inhibitory substances by repeated washing of the beads after capture without decreasing the analytical sensitivity, thereby allowing adaptation of the test to an automated high-throughput platform. However, the failure rate of 6.8% for the peptide-mediated PCR assay should be reduced further before its widespread use in routine diagnostic laboratories.
Finally, the diagnostic applicability of the peptide-mediated capture PCR was evaluated by testing bulk milk samples from 423 dairy herds for M. avium subsp. paratuberculosis. The finding that 23 of the samples were PCR positive documents that the analytical sensitivity of the test is sufficient to detect the pathogen in a substantial number of bulk milk samples under field conditions.
In summary, we developed and demonstrated the practical application of a peptide-mediated capture PCR for the detection of M. avium subsp. paratuberculosis in bulk milk samples obtained from dairy herds. The method relies solely on synthetic reagents, performs well under routine laboratory conditions, and is adaptable to standard laboratory automation. Furthermore, in contrast to antibody-based methods, peptide-mediated capture cannot be inhibited by circulating antibodies present in milk and therefore might present a powerful tool for large-scale paratuberculosis control programs.
Acknowledgments
This work was supported by the FEI (Forschungsverbund der Ernaehrungsindustrie E.V., Bonn, Germany), the AiF, and the Ministry of Economics and Labor under project no. AiF-FV 13585 N. J.H. is a fellow of the Georg-Christoph-Lichtenberg scholarship of the Ministry for Science and Culture of Lower Saxony, Hannover, Germany. K.D. was supported by EU grant QLK 2-CT-2001-01420.
REFERENCES
- 1.Amano, A., T. Nakamura, S. Kimura, I. Morisaki, I. Nakagawa, S. Kawabata, and S. Hamada. 1999. Molecular interactions of Porphyromonas gingivalis fimbriae with host proteins: kinetic analyses based on surface plasmon resonance. Infect. Immun. 67:2399-2405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Benedictus, G., and C. J. Kalis. 2003. Paratuberculosis: eradication, control and diagnostic methods. Acta Vet. Scand. 44:231-241. [PubMed] [Google Scholar]
- 3.Beyerbach, M., T. Rehm, L. Kreienbrock, and G. F. Gerlach. 2001. Eradication of paratuberculosis in dairy herds: determination of the initial herd prevalence and modelling of prevalence development. Dtsch. Tierarztl. Wochenschr. 108:291-296. [PubMed] [Google Scholar]
- 4.Birkness, K. A., W. E. Swords, P. H. Huang, E. H. White, C. S. Dezzutti, R. B. Lal, and F. D. Quinn. 1999. Observed differences in virulence-associated phenotypes between a human clinical isolate and a veterinary isolate of Mycobacterium avium. Infect. Immun. 67:4895-4901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bitsch, V., and L. Ronsholt. 1995. Control of bovine viral diarrhea virus infection without vaccines. Vet. Clin. N. Am. Food Anim. Pract. 11:627-640. [DOI] [PubMed] [Google Scholar]
- 6.Boeddinghaus, B., T. Rogall, T. Flohr, H. Blocker, and E. C. Boettger. 1990. Detection and identification of mycobacteria by amplification of rRNA. J. Clin. Microbiol. 28:1751-1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chiodini, R. J., H. J. Van Kruiningen, and R. S. Merkal. 1984. Ruminant paratuberculosis (Johne's disease): the current status and future prospects. Cornell Vet. 74:218-262. [PubMed] [Google Scholar]
- 8.Clarke, C. J. 1997. The pathology and pathogenesis of paratuberculosis in ruminants and other species. J. Comp. Pathol. 116:217-261. [DOI] [PubMed] [Google Scholar]
- 9.Cousins, D. V., R. Whittington, I. Marsh, A. Masters, R. J. Evans, and P. Kluver. 1999. Mycobacteria distinct from Mycobacterium avium subsp. paratuberculosis isolated from the faeces of ruminants possess IS900-like sequences detectable by IS900 polymerase chain reaction: implications for diagnosis. Mol. Cell. Probes 13:431-442. [DOI] [PubMed] [Google Scholar]
- 10.Doran, T. J., J. K. Davies, A. J. Radford, and A. L. Hodgson. 1994. Putative functional domain within ORF2 on the Mycobacterium insertion sequences IS900 and IS902. Immunol. Cell Biol. 72:427-434. [DOI] [PubMed] [Google Scholar]
- 11.Einhauer, A., and A. Jungbauer. 2001. Affinity of the monoclonal antibody M1 directed against the FLAG peptide. J. Chromatogr. A 921:25-30. [DOI] [PubMed] [Google Scholar]
- 12.Englund, S., G. Bolske, and K. E. Johansson. 2002. An IS900-like sequence found in a Mycobacterium sp. other than Mycobacterium avium subsp. paratuberculosis. FEMS Microbiol. Lett. 209:267-271. [DOI] [PubMed] [Google Scholar]
- 13.Fagerstam, L. G., A. Frostell-Karlsson, R. Karlsson, B. Persson, and I. Ronnberg. 1992. Biospecific interaction analysis using surface plasmon resonance detection applied to kinetic, binding site and concentration analysis. J. Chromatogr. 597:397-410. [DOI] [PubMed] [Google Scholar]
- 14.Garcia-Ojeda, P. A., S. Hardy, S. Kozlowski, K. E. Stein, and I. M. Feavers. 2004. Surface plasmon resonance analysis of antipolysaccharide antibody specificity: responses to meningococcal group C conjugate vaccines and bacteria. Infect. Immun. 72:3451-3460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gerlach, G. F., C. Anderson, A. A. Potter, S. Klashinsky, and P. J. Willson. 1992. Cloning and expression of a transferrin-binding protein from Actinobacillus pleuropneumoniae. Infect. Immun. 60:892-898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Grant, I. R., H. J. Ball, and M. T. Rowe. 1998. Isolation of Mycobacterium paratuberculosis from milk by immunomagnetic separation. Appl. Environ. Microbiol. 64:3153-3158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Grant, I. R., C. M. Pope, L. M. O'Riordan, H. J. Ball, and M. T. Rowe. 2000. Improved detection of Mycobacterium avium subsp. paratuberculosis in milk by immunomagnetic PCR. Vet. Microbiol. 77:369-378. [DOI] [PubMed] [Google Scholar]
- 18.Guerrero, C., C. Bernasconi, D. Burki, T. Bodmer, and A. Telenti. 1995. A novel insertion element from Mycobacterium avium, IS1245, is a specific target for analysis of strain relatedness. J. Clin. Microbiol. 33:304-307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hamada, T., M. Kawashima, H. Watanabe, J. Tagami, and H. Senpuku. 2004. Molecular interactions of surface protein peptides of Streptococcus gordonii with human salivary components. Infect. Immun. 72:4819-4826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hammer, P., C. Kiesner, H. G. Walte, K. Knappstein, and P. Teufel. 2003. Heat resistance of Mycobacterium avium subspecies paratuberculosis in raw milk tested in a pilot plant pasteurizer. Kieler Milchwirtschafl. Forsch. 54:275-303. [Google Scholar]
- 21.Harris, N. B., and R. G. Barletta. 2001. Mycobacterium avium subsp. paratuberculosis in veterinary medicine. Clin. Microbiol. Rev. 14:489-512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hendrick, S. H., T. E. Duffield, D. E. Kelton, K. E. Leslie, K. D. Lissemore, and M. Archambault. 2005. Evaluation of enzyme-linked immunosorbent assays performed on milk and serum samples for detection of paratuberculosis in lactating dairy cows. J. Am. Vet. Med. Assoc. 226:424-428. [DOI] [PubMed] [Google Scholar]
- 23.Hoorfar, J., B. Malorny, A. Abdulmawjood, N. Cook, M. Wagner, and P. Fach. 2004. Practical considerations in design of internal amplification controls for diagnostic PCR assays. J. Clin. Microbiol. 42:1863-1868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jark, U., I. Ringena, B. Franz, G. F. Gerlach, M. Beyerbach, and B. Franz. 1997. Development of an ELISA technique for serodiagnosis of bovine paratuberculosis. Vet. Microbiol. 57:189-198. [DOI] [PubMed] [Google Scholar]
- 25.Jayarao, B. M., and D. R. Wolfgang. 2003. Bulk-tank milk analysis. A useful tool for improving milk quality and herd udder health. Vet. Clin. N. Am. Food Anim. Pract. 19:75-92. [DOI] [PubMed] [Google Scholar]
- 26.Jubb, T. F., E. S. Sergeant, A. P. Callinan, and J. Galvin. 2004. Estimate of the sensitivity of an ELISA used to detect Johne's disease in Victorian dairy cattle herds. Aust. Vet. J. 82:569-573. [DOI] [PubMed] [Google Scholar]
- 27.Karlsson, R. 2004. SPR for molecular interaction analysis: a review of emerging application areas. J. Mol. Recognit. 17:151-161. [DOI] [PubMed] [Google Scholar]
- 28.Khare, S., T. A. Ficht, R. L. Santos, J. Romano, A. R. Ficht, S. Zhang, I. R. Grant, M. Libal, D. Hunter, and L. G. Adams. 2004. Rapid and sensitive detection of Mycobacterium avium subsp. paratuberculosis in bovine milk and feces by a combination of immunomagnetic bead separation-conventional PCR and real-time PCR. J. Clin. Microbiol. 42:1075-1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kreeger, J. M. 1991. Ruminant paratuberculosis—a century of progress and frustration. J. Vet. Diagn. Investig. 3:373-382. [DOI] [PubMed] [Google Scholar]
- 30.Kunze, Z. M., S. Wall, R. Appelberg, M. T. Silva, F. Portaels, and J. J. McFadden. 1991. IS901, a new member of a widespread class of atypical insertion sequences, is associated with pathogenicity in Mycobacterium avium. Mol. Microbiol. 5:2265-2272. [DOI] [PubMed] [Google Scholar]
- 31.Li, L., J. P. Bannantine, Q. Zhang, A. Amonsin, B. J. May, D. Alt, N. Banerji, S. Kanjilal, and V. Kapur. 2005. The complete genome sequence of Mycobacterium avium subspecies paratuberculosis. Proc. Natl. Acad. Sci. USA 102:12344-12349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Manning, E. J., and M. T. Collins. 2001. Mycobacterium avium subsp. paratuberculosis: pathogen, pathogenesis and diagnosis. Rev. Sci. Technol. 20:133-150. [DOI] [PubMed] [Google Scholar]
- 33.Marsh, I., R. Whittington, and D. Millar. 2000. Quality control and optimized procedure of hybridization capture-PCR for the identification of Mycobacterium avium subsp. paratuberculosis in faeces. Mol. Cell. Probes 14:219-232. [DOI] [PubMed] [Google Scholar]
- 34.Millar, D., J. Ford, J. Sanderson, S. Withey, M. Tizard, T. Doran, and J. Hermon-Taylor. 1996. IS900 PCR to detect Mycobacterium paratuberculosis in retail supplies of whole pasteurized cows' milk in England and Wales. Appl. Environ. Microbiol. 62:3446-3452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mozsolits, H., T. H. Lee, A. H. Clayton, W. H. Sawyer, and M. I. Aguilar. 2004. The membrane-binding properties of a class A amphipathic peptide. Eur. Biophys. J. 33:98-108. [DOI] [PubMed] [Google Scholar]
- 36.Nielsen, S. S., C. Gronbaek, J. F. Agger, and H. Houe. 2002. Maximum-likelihood estimation of sensitivity and specificity of ELISAs and faecal culture for diagnosis of paratuberculosis. Prev. Vet. Med. 53:191-204. [DOI] [PubMed] [Google Scholar]
- 37.Olsen, I., G. Sigurgardottir, and B. Djonne. 2002. Paratuberculosis with special reference to cattle. Vet. Q. 24:12-28. [DOI] [PubMed] [Google Scholar]
- 38.O'Mahony, J., and C. Hill. 2004. Rapid real-time PCR assay for detection and quantitation of Mycobacterium avium subsp. paratuberculosis DNA in artificially contaminated milk. Appl. Environ. Microbiol. 70:4561-4568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Raizman, E. A., S. J. Wells, S. M. Godden, R. F. Bey, M. J. Oakes, D. C. Bentley, and K. E. Olsen. 2004. The distribution of Mycobacterium avium ssp. paratuberculosis in the environment surrounding Minnesota dairy farms. J. Dairy Sci. 87:2959-2966. [DOI] [PubMed] [Google Scholar]
- 40.Reichel, M. P., R. Kittelberger, M. E. Penrose, R. M. Meynell, D. Cousins, T. Ellis, L. M. Mutharia, E. A. Sugden, A. H. Johns, and G. W. de Lisle. 1999. Comparison of serological tests and faecal culture for the detection of Mycobacterium avium subsp. paratuberculosis infection in cattle and analysis of the antigens involved. Vet. Microbiol. 66:135-150. [DOI] [PubMed] [Google Scholar]
- 41.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 42.Semret, M., D. C. Alexander, C. Y. Turenne, P. de Haas, P. Overduin, D. van Soolingen, D. Cousins, and M. A. Behr. 2005. Genomic polymorphisms for Mycobacterium avium subsp. paratuberculosis diagnostics. J. Clin. Microbiol. 43:3704-3712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shin, S. J., Y. F. Chang, C. Huang, J. Zhu, L. Huang, H. S. Yoo, K. S. Shin, S. Stehman, S. J. Shin, and A. Torres. 2004. Development of a polymerase chain reaction test to confirm Mycobacterium avium subsp. paratuberculosis in culture. J. Vet. Diagn. Investig. 16:116-120. [DOI] [PubMed] [Google Scholar]
- 44.Stabel, J. R., S. J. Wells, and B. A. Wagner. 2002. Relationships between fecal culture, ELISA, and bulk tank milk test results for Johne's disease in US dairy herds. J. Dairy Sci. 85:525-531. [DOI] [PubMed] [Google Scholar]
- 45.Stevenson, K., and J. M. Sharp. 1997. The contribution of molecular biology to Mycobacterium avium subspecies paratuberculosis research. Vet. J. 153:269-286. [DOI] [PubMed] [Google Scholar]
- 46.Stratmann, J., B. Strommenger, R. Goethe, K. Dohmann, G. F. Gerlach, K. Stevenson, L. L. Li, Q. Zhang, V. Kapur, and T. J. Bull. 2004. A 38-kilobase pathogenicity island specific for Mycobacterium avium subsp. paratuberculosis encodes cell surface proteins expressed in the host. Infect. Immun. 72:1265-1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stratmann, J., B. Strommenger, K. Stevenson, and G. F. Gerlach. 2002. Development of a peptide-mediated capture PCR for detection of Mycobacterium avium subsp. paratuberculosis in milk. J. Clin. Microbiol. 40:4244-4250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Streeter, R. N., G. F. Hoffsis, S. Bech-Nielsen, W. P. Shulaw, and D. M. Rings. 1995. Isolation of Mycobacterium paratuberculosis from colostrum and milk of subclinically infected cows. Am. J. Vet. Res. 56:1322-1324. [PubMed] [Google Scholar]
- 49.Strommenger, B., K. Stevenson, and G. F. Gerlach. 2001. Isolation and diagnostic potential of ISMav2, a novel insertion sequence-like element from Mycobacterium avium subspecies paratuberculosis. FEMS Microbiol. Lett. 196:31-37. [DOI] [PubMed] [Google Scholar]
- 50.Taylor, T. K., C. R. Wilks, and D. S. McQueen. 1981. Isolation of Mycobacterium paratuberculosis from the milk of a cow with Johne's disease. Vet. Rec. 109:532-533. [PubMed] [Google Scholar]
- 51.White, W. B., D. L. Whipple, J. R. Stabel, and C. A. Bolin. 1994. Comparison of cellular and extracellular proteins expressed by various isolates of Mycobacterium paratuberculosis and other mycobacterial species. Am. J. Vet. Res. 55:1399-1405. [PubMed] [Google Scholar]
- 52.Whittington, R. J., D. J. Marshall, P. J. Nicholls, I. B. Marsh, and L. A. Reddacliff. 2004. Survival and dormancy of Mycobacterium avium subsp. paratuberculosis in the environment. Appl. Environ. Microbiol. 70:2989-3004. [DOI] [PMC free article] [PubMed] [Google Scholar]