Abstract
Marine coccolithophorid algae are thought to play a significant role in carbon cycling due to their ability to incorporate dissolved inorganic carbon (DIC) into both calcite and photosynthetic products. Among coccolithophorids, Emiliania huxleyi is the most prolific, forming massive blooms that affect the global environment. In addition to its ecological importance, the elaborate calcite structures (coccoliths) are being investigated for the design of potential materials for science and biotechnological devices. To date, most of the research focus in this organism has involved the partitioning of DIC between calcification and photosynthesis, primarily using measurements of an external versus internal carbonic anhydrase (CA) activity under defined conditions. The actual genes, proteins, and pathways employed in these processes have not been identified and characterized (see the work of Quinn et al. in this issue [P. Quinn, R. M. Bowers, X. Zhang, T. M. Wahlund, M. A. Fanelli, D. Olszova, and B. A. Read, Appl. Environ. Microbiol. 72:5512-5526, 2006]). In this study, the cloning and preliminary characterization of two genetically distinct carbonic anhydrase cDNAs are described. Phylogenetic analysis indicated that these two genes belonged to the gamma (γ-EhCA2) and delta (δ-EhCA1) classes of carbonic anhydrases. The deduced amino acid sequence of δ-EhCA1 revealed that it encodes a protein of 702 amino acids (aa) (ca. 77.3 kDa), with a transmembrane N-terminal region of 373 aa and an in-frame C-terminal open reading frame of 329 aa that defines the CA region. The γ-EhCA2 protein was 235 aa in length (ca. 24.9 kDa) and was successfully expressed in Escherichia coli BL21(DE3) and purified as an active recombinant CA. The expression levels of each transcript from quantitative reverse transcription-PCR experiments under bicarbonate limitation and over a 24-h time course suggest that these isozymes perform different functions in E. huxleyi.
Single-celled marine algae fix inorganic carbon via the Calvin-Benson-Bassham cycle, resulting in the formation of 2 mol of phosphoglyceric acid from CO2 and its five-carbon acceptor. The enzyme responsible for this reaction, ribulose-1,5-bisphosphate carboxylase/oxygenase (RubisCO), requires CO2 for carbon fixation, and kinetic analyses of these enzymes from several photosynthetic microorganisms have shown that RubisCO enzymes have poor binding affinity for this substrate. The predominant form of dissolved inorganic carbon (DIC) in the oceans is bicarbonate (∼2 mM), and concentrations of aqueous CO2 are very low (10 μM), well below the half-saturation constants of most marine algal RubisCO enzymes (6). This requires that unicellular algae increase intracellular DIC levels either via the direct transport of HCO3− or by the activity of an external carbonic anhydrase (CA) (CAext). All carbon-concentrating mechanisms (CCMs) described to date in marine phytoplankton involve the zinc metalloenzyme carbonic anhydrase; therefore, CO2 acquisition via these enzymes is dependent upon the availability of trace metals (Zn, Co, and Cd), the concentrations of which are extremely low (nanomolar to picomolar levels) in ocean surface waters. These conditions place severe constraints on CO2 availability for marine phytoplankton via CA, yet growth kinetics and global distribution data on coccolithophorids and diatoms suggest that they are not generally CO2 limited for photosynthesis under most environmental conditions. Considering the low concentrations and dramatic fluctuations in these trace metals, it is logical to envision scenarios under which CA activity is repressed and an alternative CCM mechanism is induced, such as the CCM pathways observed in C4 and Crassulacean acid metabolism (CAM) plants. An example of such a mechanism was recently described for the marine diatom Thalassiosira weissflogii. The results of that elegant study provided solid evidence for C4 photosynthesis in a unicellular alga (26). Another study showed a similar C4 mechanism within a single photosynthetic cell in a terrestrial plant (44), and thus, the argument requiring a Kranz anatomy for C4 photosynthesis must be reevaluated as we learn more about the molecular biology of carbon metabolism in marine microalgae.
The calcifying marine alga Emiliania huxleyi incorporates inorganic carbon into both organic and biomineralized products under environmental conditions that remain to be elucidated. This process occurs in a specialized structure, called the coccolith vesicle, and is summarized in the following equation: Ca2+ + 2 HCO3→CaCO3 + CO2 + H+.
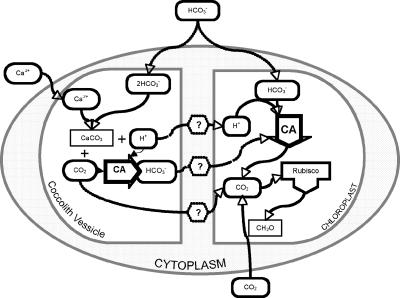
Calcification in E. huxleyi has been postulated to be the mechanism for generating additional CO2 for RubisCO, with the argument stating that this ability would eliminate the need for a traditional CCM (Fig. 1). However, despite numerous physiological studies, data confirming this hypothesis have not been obtained (4, 8, 23, 25, 28). Most models describing the intracellular conversion of HCO3− to CO2 in E. huxleyi have historically invoked calcification as the CO2 source for RubisCO (8, 33). However, the data from these studies have generated a number of conflicting scenarios, none of which adequately support or refute this hypothesis. In addition, a recent study investigating Zn-HCO3− colimitation in E. huxleyi was not able to demonstrate that E. huxleyi had a CCM by employing growth kinetic experiments and statistical analysis methodologies as the primary research tools. Moreover, the results of that study also generated additional conflicting and inconsistent data that did not resolve the CCM issue in E. huxleyi (9). Our working hypothesis is that the presence of a CA in the coccolith vesicle (CV), if confirmed, might resolve the questions of how the CO2 by-product of the calcification reaction might be available for photosynthesis (Fig. 1). A CV-located CA would recapture the CO2 produced from calcification, converting it to HCO3−. Transport of this HCO3− out of the CV would serve to increase intracellular DIC, which may be available for photosynthesis by transport into the chloroplast (Fig. 1).
FIG. 1.
Schematic representation of hypothesized relationships between photosynthesis and calcification in coccolithophorid algae. Previous models proposed that the CO2 and H+ by-products of calcification in the CV could serve to enhance photosynthesis; however, the mechanisms by which this could occur have not been demonstrated. If confirmed, a CV-located CA could resolve these questions by converting the CO2 released in the calcification reaction to HCO3− for transport out of the CV, thus increasing the amount of intracellular DIC for photosynthesis.
The use of molecular and genetic approaches to study inorganic carbon metabolism and CCMs in other unicellular freshwater and marine algae has revealed an incredible diversity of carbonic anhydrases representing all known classes of these enzymes (12, 15, 18, 19, 30, 35, 39, 43). For example, the green alga Chlamydomonas reinhardtii possesses six CAs representing two classes (α and β) with distinct cellular locations: the periplasm (Cah1 and Cah2), thylakoid lumen (Cah3 and Cah6), and mitochondria (ca1 and ca2) (19). Two genetically distinct CAs have been identified in the diatom T. weissflogii, and a novel cadmium-dependent enzyme has recently been purified and characterized from this same organism (15). The amino acid sequence of this Cd-dependent CA is distinct from all other CAs and has been proposed as yet another novel class, the ζ-carbonic anhydrase (15). These studies demonstrate that there are serious limitations in relying exclusively on CAext versus internal CA (CAint) enzyme activities as a tool for addressing questions regarding the biology and ecology of coccolithophorids. This is especially important considering the essential metabolic roles that CA enzymes have been shown to play in central and intermediary carbon metabolism in organisms from virtually all three domains of life (e.g., photosynthesis, respiration, ion transport, acid-base balance, and biomineralization).
In order to investigate the question of inorganic carbon acquisition and metabolism in E. huxleyi, our laboratory is utilizing molecular tools and employing genetic approaches to identify and characterize genes and proteins involved in these processes. Despite the significant role that these algae are thought to play in the global carbon cycle, other than the use of RubisCO as a phylogenetic marker, not a single gene or protein involved in carbon acquisition and metabolism has been cloned and characterized in these organisms. A number of studies measuring the temporal variability in RubisCO (rbcL) mRNA in mixed marine phytoplankton communities have been described and have determined that this transcript had a diel rhythm of expression (47, 50, 51). The first species-specific evaluation of RubisCO gene expression in a coccolithophorid was reported recently and revealed a diel expression pattern for this transcript (51). In this study, we report the first identification and initial characterization of two genetically distinct carbonic anhydrases in the marine coccolithophorid alga E. huxleyi that belong to the γ and δ classes of carbonic anhydrases (γ-EhCA2 and δ-EhCA1). Preliminary data on the expression of these isozymes under bicarbonate limitation conditions and over a 24-h time course using real-time reverse transcription (RT)-PCR suggest that these enzymes play very different roles during inorganic carbon acquisition in this organism. In addition, these transcripts display a difference in temporal expression, with evidence of a diel, or perhaps circadian, rhythm shown for γ-EhCA2 but not for δ-EhCA1. We also describe the heterologous expression and purification of active recombinant E. huxleyi γ-EhCA2 from Escherichia coli.
MATERIALS AND METHODS
Strains, plasmids, and growth conditions.
E. huxleyi strain CCMP 2090 used in this study was obtained from the Provasoli-Guillard National Center for the Culture of Marine Phytoplankton (CCMP). A modification of f/2 artificial seawater medium employed in this study (14), called “phosphate-replete f/2,” was prepared by the addition of the following components (per liter of double-distilled water): 400 mM NaCl, 10 mM KCl, 20 mM MgSO4 · 7H2O, 20 mM MgCl2 · 6H2O, 7.5 mM CaCl2 · 2H2O, 0.4 mM HBO3, 0.88 mM NaNO3, 0.036 mM NaH2PO4 · H2O, trace elements (76.5 nM ZnSO4 · 7H2O, 39 nM CuSO4 · 5H2O, 42 nM CoCl2 · 6H2O, 910 nM MnCl2 · 4H2O, 26 nM Na2MoO4 · 2H2O, 5.8 μM FeCl2 · 6 H2O, 11.7 μM Na2EDTA · 2H2O), and vitamin solution (50 μg biotin, 50 μg vitamin B12, 100 ng thiamine-HCl). Next, 10 mM Tris buffer was added, and the medium pH was adjusted to 7.8 with concentrated HCl prior to autoclaving. Medium pH was maintained with this amount of Tris regardless of the HCO3− levels employed in this study. Filter-sterilized bicarbonate was added to the cooled medium following autoclaving as described below for the various experiments. Bicarbonate-replete medium contained 2.2 mM NaHCO3. Strain 2090 is an axenic isolate from CCMP 1516 and does not calcify under any conditions tested thus far. Cells from late log to early stationary phase (∼2 × 106 cells · ml−1) were inoculated (1 × 10−2 dilution) into 4-liter flasks containing 1 liter of medium and incubated at 18 to 20°C under cool white fluorescent light (660 μm · m−2 · s−2) with a discontinuous photoperiod (12 h light and 12 h dark).
E. coli strains Top10 and BL21(DE3) possessing recombinant plasmids were grown in LB medium (29) with ampicillin (100 μg/ml). Recombinant plasmids were routinely maintained in Top10 and were transformed into BL21(DE3), lysed to initiate each expression experiment, as recommended by the manufacturer's protocol (Invitrogen, Inc.).
RNA isolation, cDNA synthesis, and qRT-PCR.
RNA was isolated from 500 ml early-stationary-phase (9 to 10 days) cells as previously described (20). First-strand cDNA synthesis was performed using an Omniscript cDNA synthesis kit (QIAGEN Inc., Valencia, CA). For cDNA synthesis, a 20-μl reaction mixture contained 2 μg template RNA, 1× RT buffer, 0.5 mM deoxynucleoside triphosphate, 1 μM oligo(dT) primer, 10 units of RNase inhibitor, and 4 units of Omniscript reverse transcriptase. The reaction mixture was incubated for 60 min at 37°C. cDNA was diluted 1:100, and 5 μl of the dilution was used in a SYBR green quantitative RT (qRT)-PCR.
qRT-PCR was carried out using SYBR green chemistry for amplicon detection. The SYBR green assays were performed using the iCycler iQ system (Bio-Rad). Amplification of target genes was performed in triplicate in a 96-well plate. Each 25-μl reaction mixture contained 5 μl cDNA, 7.1 μl 2× SYBR green master mix, and 0.24 μM each forward and reverse primers. The cycling conditions for amplification included a 10-min polymerase activation step at 95°C followed by 40 cycles of 95°C for 10 s, 60°C for 30 s, and 82°C for 30 s. Fluorescence was measured at 82°C during the final 30 s of each cycle. After PCR amplification, melting curves were generated by denaturing the sample for 1 min at 95°C, cooling to 55°C for 1 min, and then ramping the temperature 0.5°C every 10 s beginning at 55°C until a final temperature of 95°C was reached. Serial dilutions (from 10 pg to 1 fg) of the cDNA library plasmids pSPORT-ehCA1 and pMAB-ehCA2 were used as templates to generate standard curves and for quantifying the transcript copy number. The primers used for qRT-PCR were as follows: 5′-GTAGCGGGAGCACATTTCG-3′ (forward) and 5′-GGATGGACGGGAAGGACAAG-3′ (reverse) for the EhCA1 gene and 5′-TTTGGTCGCTGGTCGAACAG-3′ (forward) and 5′-CGCAGGACAACTGGATTTTCCA-3′ (reverse) for the EhCA2 gene. Standards of serially diluted plasmids were run with each of the experimental samples to determine the copy number. To monitor for potential contamination, all reactions were run with no-template controls. Expression levels for each transcript were expressed as copy numbers calculated from the standard curve.
Plasmid constructions and expression of E. huxleyi recombinant carbonic anhydrase in E. coli cells.
The E. huxleyi gamma carbonic anhydrase gene identified from our f/50 cDNA library (45, 46) was subcloned into the expression vector pET100-TOPO. The cloning strategy involved two steps. The γ-ehCA2 open reading frame (ORF) was amplified from cDNA library clone pMAB-ehCA2 by PCR using the following primers: 5′-CACCATGAAGCGGGTGCTCGTCGG-3′ (forward) and 5′-CTAGATCTTGCCGGCGGCC-3′ (reverse). The forward primer contained four vector nucleotides (underlined) on the 5′ end for directional cloning into pET100. The δ-EhCA1 gene ORF was amplified from cDNA library clone pSPORT-ehCA1with the following primers: 5′-CACCATGTCGCAGGCGGATTGGC-3′ (forward) and 5′-TCACTTGCGGGACTGCTGG-3′ (reverse). The resultant PCR products were gel extracted prior to TOPO cloning and transformed into E. coli Top10 cells (Invitrogen, Inc.), and transformants were selected on LB plates with ampicillin (100 μg/ml). Positive clones were verified by restriction enzyme digests and PCR. Sequencing was performed (Retrogen, Inc., San Diego, CA) to confirm that proteins were in the proper reading frame in the recombinant plasmid before expression experiments were initiated. Expression experiments were initiated by transforming recombinant pET-ehCA2 and pET-ehCA1 plasmid DNA into E. coli strain BL21(DE3). A single colony from a fresh transformation reaction mixture (1- to 2-day-old platings) was picked and grown overnight in 2 ml LB (ampicillin). Cells from the culture grown overnight were inoculated into 20 ml of same medium (1:50 dilution) and grown to an optical density at 600 nm of 0.5 to 0.8. The cells were divided into two 10-ml cultures (50-ml flask), one of which was induced by the addition of IPTG (isopropyl-β-d-thiogalactopyranoside) (0.5 to 1 mM). Following an additional 3 h of growth, cells were harvested by centrifugation (5,000 × g for 10 min at 4°C), and pellets were frozen at −20°C until use.
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), Western blots, and purification of γ-EhCA2.
Cells from small-scale expression experiments were prepared for protein extractions and Western blotting in the following manner. Cell pellets from expression experiments were resuspended in extraction buffer (50 mM Tris, pH 7.8, 150 mM NaCl, 5 mM BME [β-mercaptoethanol], 1 mM phenylmethylsulfonyl fluoride) and lysed by sonication at 0°C with three 10- to 15-s bursts followed by 30-s cooling periods. Following centrifugation (10,000 × g for 10 min at 4°C), supernatant and pellet fractions were run on 4 to 12% gradient gels (NuPAGE Tris-acetate; Invitrogen, Inc.). Protein bands were detected using the Simple Blue staining protocol according to the instructions of the manufacturer (Invitrogen, Inc.), and relative size was determined using SeeBlue molecular weight standards (Invitrogen, Inc.). Confirmation of recombinant γ-EhCA2 expression was done by Western blotting employing the anti-Xpress antibody directed against the N-terminal Xpress epitope tag encoded on the pET100 vector according to the manufacturer's instructions (Invitrogen, Inc.). For purification, 500 ml cells from cultures induced for 3 h were harvested, and His-tagged recombinant protein was obtained using the Ni-nitrilotriacetic acid purification system kit according to the instructions of manufacturer (Invitrogen, Inc., Carlsbad, CA).
Assays of recombinant γ-EhCA2 enzyme activity.
Assays of recombinant γ-EhCA2 activities were measured electrometrically using the Wilbur-Anderson method (48). Extract containing various amounts of recombinant γ-EhCA2 protein was added to 800 μl of buffer (50 mM Tris-Cl, pH 8.3), and the reaction was initiated with the addition of 600 μl of ice-cold CO2-saturated water. The pH change resulting from CO2 hydration was measured by a combination microelectrode (Microelectrodes, Inc.). One Wilbur-Anderson unit (WAU) of activity is defined as T0 − T)/T, where T0 (uncatalyzed reaction) and T (catalyzed reaction) are recorded as the time required for the pH to drop from 8.3 to 6.3. Protein was determined using the Bio-Rad microassay (Bio-Rad). Specific activity was expressed as WAU/milligram of protein.
Materials.
All chemicals were of reagent grade and were purchased from Sigma Chemical or Fisher Scientific. Highly purified bovine carbonic anhydrase was purchased from Sigma. Oligonucleotide primers were obtained from Invitrogen, and sequencing was performed at Retrogen, Inc. (San Diego, CA) and the Microchemical Core Facility at San Diego State University.
Nulceotide sequence accession numbers.
The sequences described in this article have been deposited in GenBank under accession numbers DQ644550 (δ-EhCA1 gene) and DQ644551 (γ-EhCA2 gene).
RESULTS
Analysis of gene expression patterns in batch culture using real-time RT-PCR.
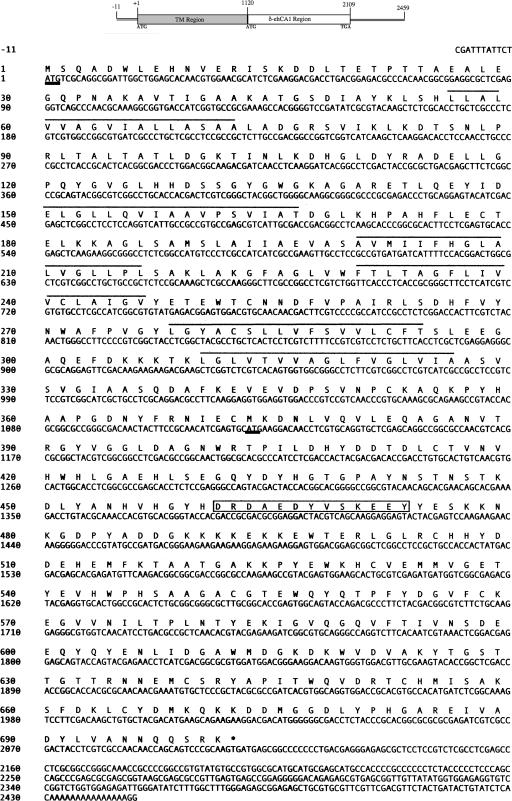
Our initial efforts to determine if the E. huxleyi cDNA sequences encoded functional carbonic anhydrase enzymes required careful analysis of their deduced amino acid sequences. From our expressed sequence tag sequence analysis of cDNAs from noncalcifying cells of E. huxleyi (46), we identified a cDNA that showed significant sequence identity (BLAST e value, 10−70) to the zinc-dependent intracellular carbonic anhydrase (TWCA1) from the marine diatom Thalassiosira weissflogii (27). Full-length sequencing of this cDNA (designated EhCA1) revealed that it was composed of an 11-bp 5′ untranslated region (UTR), an open reading frame of 2,109 bp, and a 336-bp 3′ UTR with a potential 15-bp poly(A) tail (Fig. 2). The deduced amino acid sequence of the EhCA1 gene was predicted to encode a protein of 702 amino acids (aa) in length, with a calculated molecular mass of 77.3 kDa and a theoretical pI of 5.40.
FIG. 2.
Nucleotide sequence and deduced amino acid sequence of the 2,456-bp δ-EhCA1 gene cDNA from E. huxleyi. A diagram of the cDNA clone showing the relative positions of the two proposed ORFs is shown above the sequence. The putative translational start of the 2,109-bp ORF is underlined in boldface type at bp 1, and the translation termination codon (TGA) is indicated with an asterisk. The in-frame ATG defining the start of the CA region of the transcript is underlined in boldface type (M 374). The putative six transmembrane domains are delineated with lines above the involved residues.
Analysis of over 3,000 expressed sequence tags from a cDNA library constructed from cells grown under calcifying conditions (45) identified a second carbonic anhydrase in E. huxleyi that showed significant similarity (BLAST e value, 10−49) to members of the gamma class of CA enzymes. Full-length sequencing analysis of this cDNA identified an ORF of 708 bp encoding a putative polypeptide of 235 amino acids, with a calculated molecular mass of 24.9 kDa and a theoretical pI of 7.68. The cDNA transcript was 1,125 bp long, with 35 bp of sequence upstream of the putative start of the ORF and 382 bp of downstream 3′ UTR sequence following the stop codon, which includes a poly(A) tail of 53 bp (Fig. 3).
FIG. 3.
Nucleotide sequence and deduced amino acid sequence of the 1,125-bp γ-EhCA2 gene from E. huxleyi. The start codon (Met 1) is underlined, and the termination codon is indicated with an asterisk.
In order to begin an analysis of the metabolic roles played by δ-EhCA1 and γ-EhCA2 in E. huxleyi, we decided to evaluate their expression under noncalcifying (optimal growth) conditions and also in response to HCO3− limitation using qRT-PCR. Real-time RT-PCR primers were designed for each CA ORF using Primer Express software. These experiments were designed to establish baseline data for the expression of genetically distinct CA enzymes in response to HCO3− at a constant level of CO2 (ambient, ∼380 ppm). To eliminate any effects related to calcification and to focus more explicitly on the roles that these isozymes play during photosynthesis and central and intermediary carbon metabolism, we employed the axenic, noncalcifying isolate of CCMP 1516, strain CCMP 2090. Strain 2090 cells were transferred from late log to early stationary phase (∼1 × 106 to 3 × 106 cells/ml) into phosphate-replete (f/2) medium under their respective HCO3− levels or ambient CO2. After more than 20 generations, total RNA was extracted. The pH of the Tris-buffered medium used in experimental cultures did not appreciably change by the end of each experiment.
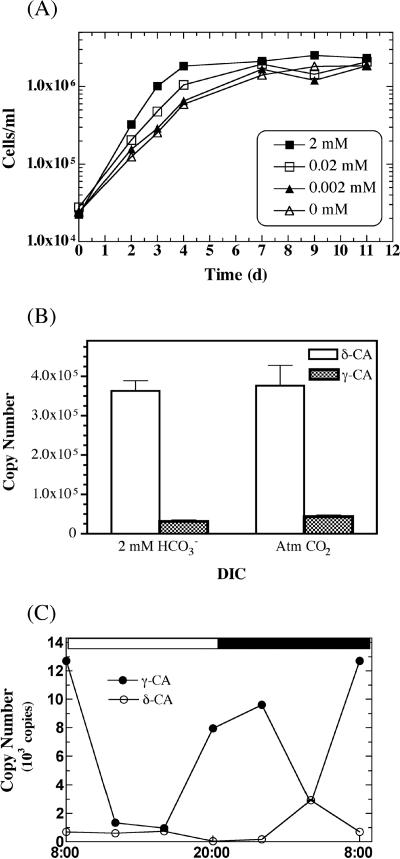
The growth rates of CCMP 2090 were slightly lower in cells grown at 0.002 mM HCO3− and at ambient CO2 than in cells grown at 2 mM HCO3−; however, the overall cell yields were the same by days 8 to 9 (Fig. 4A). After 30 generations, the growth rate of cells at ambient CO2 levels, in the absence of HCO3−, were similar to those of cells grown at 2 mM HCO3− (generation time, ∼21 h), indicating an adaptive response of E. huxleyi cells over time (data not shown). This adaptation suggests that the extended incubation with atmospheric CO2 as the sole carbon source may induce higher levels of a periplasmic (“external”) CA to increase the amount of bicarbonate within the cell for transport to the chloroplast. The lack of any significant effect of bicarbonate limitation on the growth rate was also reflected in the transcript levels of both isozymes in response to the HCO3− concentration. Figure 4B shows the relative transcript abundances of the δ-EhCA1 and γ-EhCA2 genes at 2 mM HCO3− versus HCO3− minus (ambient atmospheric CO2, ∼380 ppm). Although the copy number of each isozyme was similar regardless of the inorganic carbon source, the expression level of the δ-EhCA1 gene transcript was upregulated by ca. 12- and 9-fold over that of the γ-EhCA2 gene at 2 mM and 0 mM HCO3−, respectively (Fig. 4B). Based upon previous physiological studies describing CAext and CAint enzyme activities, we would have predicted no difference in either CAext or CAint enzyme activity in response to HCO3− limitation. However, this was not the case when the expression levels of the two genetically, and perhaps functionally, distinct CAs shown here were measured. Although the levels of HCO3− employed in this study did not result in differences in expression levels of either δ-EhCA1 or γ-EhCA2 (i.e., same copy number with or without HCO3−), the transcript levels between these isozymes were significantly different from one another. These differences could not have been detected by using external and internal CA inhibitor techniques.
FIG. 4.
Expression of E. huxleyi CCMP 2090 δ-EhCA1 gene versus the γ-EhCA2 gene using qRT-PCR. (A) Growth curves of E. huxleyi CCMP 2090 cultures grown under various DIC levels. (B) Copy numbers of the δ-EhCA1 and γ-EhCA2 genes from early-stationary-phase cells grown with HCO3− versus ambient CO2 as the carbon source. (C) Temporal expression of mid-log-phase cells showing the relative copy number of each transcript.
The significantly higher transcript levels of the δ-EhCA1 gene in cells during late log phase to stationary phase compared to the levels of γ-EhCA2 led us to investigate whether there was a difference in the temporal regulation of the expression of these isozymes. Consequently, we monitored their relative expression levels over a 24-h period, again under noncalcifying conditions. Cells were transferred into f/2 medium containing 2 mM HCO3− and incubated under a 12-h light/dark cycle to mid-log to late log phase (7 days; ∼8 × 105 cells/ml). Cells were harvested at 4-h intervals over a 24-h period, and RNA was extracted immediately at each time point and stored at −70°C prior to cDNA synthesis and qRT-PCR. The most striking observation from these data was the expression pattern of the γ-EhCA2, which showed the highest transcript levels during the dark period and the lowest transcript levels during daylight hours (Fig. 4C). The expression level of the γ-EhCA2 gene transcript began to increase after about 8 h in the light and prior to the onset of the dark phase (i.e., between 4 p.m. and 8 p.m.). In contrast, δ-EhCA1 gene expression did not show any evidence for periodicity of expression but remained relatively constant over 24 h, except for a small spike in the copy number at 20 h. In addition, the overall levels of the δ-EhCA1 gene transcript were much lower in these log-phase cells than those of the γ-EhCA2 gene transcript. This is in contrast to the expression levels observed in stationary-phase cells, which showed significantly a higher level of expression of the δ-EhCA1 gene (Fig. 4B). However, these differences most likely reflect the cell cycle versus growth phase expression patterns, given that the average doubling time of E. huxleyi is ca. 24 h. The temporal versus spatial expression story of these two isozymes will require further experimental investigation.
Sequence analysis of δ-EhCA1 and γ-EhCA2 from E. huxleyi.
A closer look at the sequence data of the EhCA1 gene cDNA identified two reading frames that defined distinct polypeptide “regions”: an upstream (N-terminal) membrane-spanning domain (373 aa), and a downstream (C-terminal) CA-containing domain (329 aa). The putative translational ATG start codon of the transcript (Met-1) reads through to another in-frame ATG (Met-374), the latter of which defined the start of the CA ORF. The transcript terminates with a TGA stop codon directly following Lys-702 (Fig. 2), followed by 336 bp of downstream sequence. Although the presence of this clone in our cDNA library suggests that this mRNA transcript is not processed prior to translation, we do not know at this point if this is actually the case. In addition, it is also not known if this transcript encodes a single bifunctional protein or, alternatively, is posttranslationally processed into two functionally distinct proteins. Analysis of the 3′ UTR using a variety of protein prediction programs (PSORT, CHLOR P, and TARGET P) failed to identify any signal sequences targeting this protein to a specific cellular location or any transcriptional/translational regulatory sequences described in the databases.
Analysis of the deduced amino acid sequence of EhCA1 using 3D-PSSM (http://www.sbg.bio.ic.ac.uk/∼3dpssm) and PSORT (http:/psort.ims.u-tokyo.ac.jp) predicted six strong transmembrane domains within the first 373 residues of this putative protein (Fig. 2). This analysis also predicted a cytosolic location for the CA region of this protein (residues 374 to 702). Similarity searches against the membrane-spanning upstream region failed to identify any similar proteins in GenBank, so we employed the sequence-independent Propsearch program (http://www.infobiosud.cnrs.fr/SERVEUR/PRPOSEARCH) to identify putative protein family homologs. The results of this analysis indicated that EhCA1 had significant similarity (94% reliability score) to cation efflux system proteins, including the cobalt-zinc-cadmium efflux protein CzcD family (5, 13). Weaker homology (14%) to a chloride-nitrate transporter from Arabidopsis thaliana was also found. Localization prediction analysis (PSORT) failed to identify any signal sequences for targeting to the chloroplast, mitochondria, endoplasmic reticulum, nucleus, or Golgi body. However, PROSITE results indicated a high probability for the presence of several sites characteristic of calcium-binding proteins, including sulfation and phosphorylation sites, and an EF-hand calcium binding motif (residues 462 to 474) (Fig. 2).
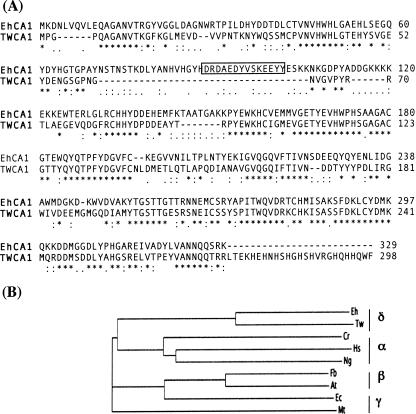
Figure 5 shows the alignment of the CA region of δ-EhCA1 with the delta class TWCA1 from the diatom T. weissflogii. The CA region of δ-EhCA1 had significant identity (51%) to TWCA1 from T. weissflogii (Fig. 5A). N-terminal sequencing of the 34-kDa enzyme purified from the diatom revealed that the first 72 residues following the Met start codon were processed to yield a 27-kDa protein. The function of this cleaved region has not been determined, and no obvious targeting or signal sequences were detected (17, 27). The percent similarity between the diatom and E. huxleyi sequences within this same region was high (∼60%), except for the presence of the EF-hand Ca2+-binding domain in the E. huxleyi sequence, which was absent in the TWCA1 sequence (Fig. 5A, boxed residues 89 to 101, and Fig. 2, boxed residues 462 to 474). Although calcium transport by CAs has been described for some eukaryotic enzymes (7), the role of this motif (if any) in the biology of E. huxleyi is presently unknown. Given the possibility that the N-terminal 373-aa polypeptide of this transcript may encode a transporter function, it will be essential to identify the cellular location of this isozyme and to determine if it is a bifunctional protein or is posttranslationally processed into distinct functional proteins. Phylogenetic analysis of the δ-EhCA1 protein against representative CAs from all known classes showed the E. huxleyi CA sequence to be distinctly different from alpha, beta, and gamma class carbonic anhydrases. The significant sequence identity of EhCA1 with TWCA1 from T. weissflogii indicated a common evolutionary history and supported its classification as the second member of the recently described delta class of CAs (Fig. 5B).
FIG. 5.
(A) Comparison of the deduced amino acid sequence of the CA region of the δ-EhCA1 gene with TWCA1 from the diatom T. weissflogii. The sequences were aligned using ClustalW. Asterisk, identical residues; colon, conserved substitutions; period, semiconserved substitutions. The sequence numbers correspond to the initiating Met from TWCA1 (Met-374 δ-EhCA1). (B) Phylogenetic tree of δ-EhCA1 with representative enzymes from the other classes of CAs: gamma class, Thalassiosira weissflogii (Tw) (GenBank accession no. AAQ56178); alpha class, Chlamydomonas reinhardtii (Cr) (accession no. AAC49983), Homo sapiens (Hs) (accession no. P00918), and Neisseria gonorrhoeae (Ng) (accession no. Q50940); beta class, Flaveria bidentis (Fb) (accession no. AAO17574) and Arabidopsis thaliana (At) (accession no. AAM61583); gamma class, E. coli (Ec) (accession no. ZP_00925010) and Methanosarcina thermophila (Mt) (accession no. 1QRGIA).
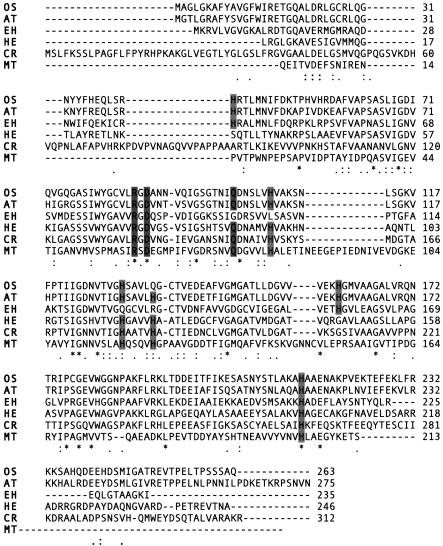
The γ-EhCA2 cDNA was also analyzed for sequence elements that might shed more light on its possible cellular location and function using a variety of tools. However, a closer look at the 5′ and 3′ UTRs using protein prediction tools (UTResource [http://bighost.area.ba.cnr.it/BIG/UTRHome/]) failed to reveal any significant signal sequences targeting the enzyme to a specific cellular location. Figure 6 shows the results from the alignment of E. huxleyi γ-EhCA2 against plant, algal, and bacterial gamma class enzymes. A BLASTP search with γ-EhCA2 revealed the highest sequence identities to γ-CA from plants, rice (Oryza japonicum [42%]), Arabidopsis (41%), and the invertebrate pathogenic nonphotosynthetic green alga Helicosporidium species (36%), with lower identities to the enzyme from Chlamydomonas (28%) and the archaeon Methanosarcina thermophila (15%) (Fig. 6). Using sequence numbering for M. thermophila, for which the crystal structure is known, several sequence features present in other γ-CAs could also be identified in the E. huxleyi sequence. The amino acid residues Arg-59, Asp-61, and Gln-75 have been strictly conserved in all prokaryotic γ-CAs identified to date. These residues were also present in the sequences from Oryza sativa and A. thaliana. However, E. huxleyi γ-EhCA2 lacked the Gln-75 residue (Fig. 4B). Of particular note, the conserved Zn ligands His-81, His-117, and His-122 present in all gamma class prokaryotic carbonic anhydrases were completely absent from the E. huxleyi enzyme. This suggests that alternative residues may coordinate the Zn or that another metal cofactor may be employed in the E. huxleyi enzyme (e.g., Co or Cd) requiring a different ligand coordination motif. It is important to note that there are relatively few reports of γ-CAs in eukaryotes, and variations in this gamma class metal ligand motif will arise as more of these enzymes are identified and characterized in eukaryotes. For example, the His-122 residue component is also absent in the rice enzyme but is present in the Arabidopsis sequence (Fig. 6). The fact that we have presented evidence to support the conclusion that γ-EhCA2 is a functional CA, based upon measurement of CA enzyme activity of the recombinant enzyme and upon qRT-PCR analysis of its transcript levels under various growth conditions (see below), indicates that we have much to learn regarding the structural and mechanistic diversity of these enzymes.
FIG. 6.
Comparison of the deduced amino acid sequence of γ-EhCA2 with sequences of γ-CAs from other selected organisms. Abbreviations and GenBank accession numbers are as follows: OS, Oryza sativa, accession no. BAD82610; AT, Arabidopsis thaliana, accession no. AAM61583; HE, Helicosporidium sp., accession no. AAU93943; CR, Chlamydomonas reinhardtii, accession no. AAR82950; MT, Methanosarcina thermophila, accession no. 1QRGIA. Sequences were aligned with ClustalW. Shaded boxes represent conserved residues present from γ-CAs that are present in or absent from γ-EhCA2.
Heterologous expression of recombinant E. huxleyi CA.
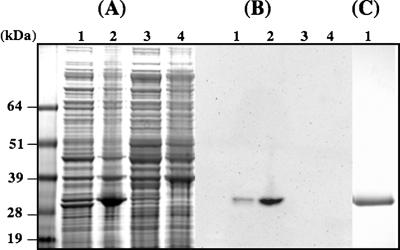
The γ-EhCA2 ORF from E. huxleyi was amplified from the cDNA library clone and directionally cloned into the pET100-TOPO vector for inducible expression and purification of recombinant protein. Expression of the protein was obtained following induction with IPTG (1 mM) for 3 h. SDS-PAGE analysis of extracts from uninduced versus induced cultures showed the presence a protein of the expected size in the induced pellet and supernatant fractions (Fig. 7A). The expected size of the recombinant γ-EhCA2 (ca. 29 kDa) was larger than the predicted polypeptide size (24.9 kDa) due to the additional vector-encoded 35 amino acids tagged onto the N terminus of the protein. Expression of γ-EhCA2 was confirmed by Western blot analysis using antibody against the Xpress epitope tag on the N terminus of the recombinant protein (Fig. 7B). Assays for recombinant CA activity were performed on crude extracts from uninduced and induced BL21(DE3) cultures. Extracts from BL21(DE3) cultures containing the pET-ehCA2 plasmid showed significant CA activity in both induced supernatant and pellet fractions (26.7- and 39-fold, respectively) compared to uninduced and control cultures (Table 1). Specific activities were normalized to BL21(DE3) cells minus the pET-ehCA2 plasmid. Levels of CA activity in uninduced supernatant fractions containing plasmids were over twofold higher than those of the control extracts, indicating a basal level of expression from the T7 promoter in the BL21(DE3) strain. Interestingly, the highest specific activities of recombinant γ-EhCA2 were found in the induced pellet fractions (specific activity, 8.1 WAU), although the enzyme would be predicted to have a cytosolic location (Table 1). The significantly higher CA enzyme activities in BL21(DE3) extracts containing recombinant γ-EhCA2 than those in BL21(DE3) extracts minus plasmid could not be attributed to a copy number effect, as pET-TOPO plasmids are of low copy number (three to four copies per cell), and expression levels in all extracts were normalized to the control CA specific activities. Although a significant proportion of the recombinant γ-EhCA2 protein appeared to be in the pellet fraction, the expression protocol was scaled up, and the recombinant protein was purified to near homogeneity under denaturing conditions (Fig. 7C). The purified protein is being used to generate antibodies for further characterization of this E. huxleyi CA. A similar strategy employed for heterologous expression of δ-EhCA1 by subcloning the entire ORF (2,109 bp) (Fig. 2) in frame into pET100-TOPO has thus far not been successful.
FIG. 7.
Expression and purification of recombinant E. huxleyi γ-EhCA2 in E. coli BL21(DE3). (A) SDS-PAGE (4 to 12%) showing proteins from induced cultures (lanes 1 and 2, soluble and pellet fractions, respectively) and from uninduced cultures (lanes 3 and 4, soluble and pellet fractions, respectively). Overexpression of recombinant γ-EhCA2 can be seen in lanes 1 and 2 (arrow). (B) Western blot of SDS gel shown in A confirming the expression of E. huxleyi γ-EhCA2 in E. coli using an antibody against the vector-encoded N-terminal Xpress epitope. Lane numbers correspond to those shown in A. (C). SDS-PAGE (4 to 12%) of recombinant E. huxleyi γ-EhCA2 purified by Ni-nitrilotriacetic acid agarose column chromatography (Invitrogen, Inc.) under denaturing conditions (lane 1).
TABLE 1.
Enzyme activities of recombinant γ-EhCA2
| BL21(DE3) strain | Protein fractiona | Sp actb (WAU/mg) | Fold changec |
|---|---|---|---|
| Without pET-ehCA2 | I-S | 0.208 | 1 |
| With pET-ehCA2 | U-S | 0.472 | 2.3 |
| U-P | NDd | ND | |
| I-S | 5.55 | 26.7 | |
| I-P | 8.10 | 39 |
I, induced; U, uninduced; S, supernatant; P, pellet.
One WAU = (T0 − T)/T, where T0 is the time for the uncatalyzed reaction and T is the time for the enzyme catalyzed reaction. Protein was determined by the BioRad assay system.
Represents the difference between the specific activity of the control strain minus the plasmid and that of the induced/uninduced recombinant strains. Specific activities represent the average data from two independent expression experiments.
ND, not detectable.
DISCUSSION
The rate-limiting step in the reduction of inorganic carbon to cell material during photosynthesis is RubisCO, and the evolutionary adaptations found in photosynthetic prokaryotes and eukaryotes to deal with this step can be summarized as two general mechanisms: (i) the evolution of a higher-affinity RubisCO (C3 plants) or (ii) the development of a CCM to increase the CO2 concentration near the active site of RubisCO (C4 plants and aquatic phototrophs). The usage of bicarbonate by algae has been shown to require an external CA and plasma membrane transporters and appears to correlate strongly with the presence of a CCM (6). In E. huxleyi, it is thought that a CA in the chloroplast generates CO2 for RubisCO, and although CA activity has been detected in chloroplasts from fractionation experiments (22), the activity was not detected during log-phase growth or in cells with low levels of calcification. Most of those physiological studies have argued for and against the operation of a CCM in E. huxleyi (21, 22, 31), based upon measurements of DIC uptake and extracellular (membrane-impermeable) and intracellular (membrane-permeable) inhibitors of carbonic anhydrase. Studies of a number of strains of coccolithophorids have indicated that photosynthetic DIC utilization values vary widely, thus making interpretations of these data difficult, and the numerous mathematical models generated to explain these relationships have not resolved this issue. In addition, the use of general CA inhibitors may also be problematic, considering that many microalgae, plants, and animals possess multiple CA isozymes, and a large number of enzymes involved in cellular metabolism require HCO3− or CO2 as a substrate, the latter of which would routinely be provided by carbonic anhydrase. Thus, the effects of these general inhibitors of CA activity on the physiology of the cell may be pleiotrophic, affecting cellular metabolism in ways not discernible using these methodologies. A particularly important (and fundamentally essential) consideration that has not been addressed in previous studies that evaluated the roles that CAs play in coccolithophorid biology is the variety of nonphotosynthetic or non-calcification-related functions that these enzymes perform in central and intermediary carbon metabolism, ion transport, pH homeostasis, and calcification.
Recombinant γ-EhCA2 was expressed in E. coli as a catalytically active enzyme, with significant activity present in both soluble and pellet fractions. Antibody is being raised against the recombinant enzyme, which will allow us to localize γ-EhCA2 and study its expression profile in E. huxleyi. This approach will complement the data obtained from qRT-PCR, allowing us to monitor the activity of specific carbonic anhydrases (and other genes) during carbon acquisition and metabolism and under calcification conditions. As this is the first molecular study of genes involved in carbon metabolism in coccolithophorids, it was important to eliminate effects due to calcification to begin to elucidate the roles of these CAs during photosynthesis and during central and intermediary carbon metabolism. In that regard, the qRT-PCR data obtained from noncalcifying, early-stationary-phase cells indicated that δ-EhCA1 and γ-EhCA2 were expressed at very different levels, which suggests different roles in cellular carbon acquisition and metabolism. Neither isozyme exhibited any difference in expression levels with decreasing HCO3− or with ambient CO2 as the sole carbon source; however, the transcript levels of δ-EhCA1 were consistently higher (ca. 12-fold) than those of γ-EhCA2 under all DIC conditions employed. Previous studies of E. huxleyi were unable to detect differences in CAext or CAint enzyme activities in E. huxleyi with bicarbonate limitation (9) or high and low partial CO2 pressure (pCO2) (28); however, the presence of multiple CA isozymes (as is the case in E. huxleyi) could certainly explain those results. We now know that, at least at the transcriptional level, these two E. huxleyi isozymes are differentially expressed with respect to one another. Based upon sequence analysis indicating that δ-EhCA1 may be a membrane-bound protein, it is tempting to speculate that higher expression levels of this enzyme in the cell may represent the “external” CA activity that has been described in the literature over the past 20 years. However, further experiments need to be employed to unequivocally determine its cellular location, and this work is in progress. It will also be important to evaluate the expression of these isozymes at low and high pCO2, as variations in pCO2 have been shown to dramatically affect CA expression in other marine algae (17, 49).
In order to obtain preliminary data on temporal variability in expression patterns of these isozymes during carbon metabolism, qRT-PCR was performed using RNA extracted from mid-log-phase cells over a 24-h period. Copy numbers of δ-EhCA1 showed a constitutive level of expression of this transcript over 24 h. This was in contrast to the results obtained with γ-EhCA2, which showed evidence of a diel (or possibly circadian) expression pattern, wherein the copy number decreased at the beginning of the light phase and began to increase prior to the start of the dark phase. It is not clear at this point whether the increase in the expression of this transcript independent of a light stimulus represents a circadian rather than a diel expression profile, as definitive experiments designed to resolve this question are in progress. The important point from these initial studies is that there was an 80% decrease in the transcript levels of γ-EhCA2 during the 12-h incubation phase in light, with levels of δ-EhCA1 showing no such pattern. Diel rhythms in the expression of carbon metabolism genes have been described for several phototrophic organisms, including RubisCO from the coccolithophorid Coccolithus pelagicus (51), and genes from other phototrophs that are regulated on a circadian clock have also been described. As mentioned above, we have employed noncalcifying cells to begin to elucidate the roles of these CAs in E. huxlyei in order to eliminate the variables from another complex process that also requires inorganic carbon (i.e., calcification). However, preliminary data from experiments currently in progress using calcifying cells indicated that transcript levels of γ-EhCA2 were upregulated about 25-fold under calcifying versus noncalcifying conditions in E. huxleyi, whereas δ-EhCA1 showed no difference in transcript levels under those same conditions (24a). It is interesting to speculate on a location of γ-EhCA2 in the coccolith vesicle based upon these preliminary qRT-PCR data, in addition to the fact that its basic pI of 7.68 would be compatible with a role in calcification, as opposed to that of the δ-EhCA1, which has an acidic pI of 5.40. The point from these initial expression studies is that without a genetic system for the manipulation of E. huxleyi, the expression levels of these two distinct CA isozymes must be carefully evaluated in response to a variety of environmental conditions that affect their expression. These include trace-metal limitation, pCO2 variation, and calcification. Experiments are in progress in our laboratories to evaluate each of these variables.
The δ-EhCA1 gene transcript was isolated from E. huxleyi as a single mRNA; however, it is not at all clear whether δ-EhCA1 functions as a single protein or is posttranslationally processed to yield two functionally distinct proteins. Sequence analysis did not reveal any potential proteolytic processing sites or signal sequences targeting it to any organelle. The upstream ORF encodes a transmembrane polypeptide of 373 aa, which suggests that it may serve a transport function, the location and identification of which remains to be determined. The downstream region of the δ-EhCA1 gene cDNA encodes a CA with significant similarity to TWCA1 from T. weissflogii and would thus be classified as the second member of the recently proposed delta class of CAs (43). In T. weissflogii, the cDNA encodes a protein of 34 kDa that is processed at the amino terminus to yield the in vivo active form of 27 kDa (27). The function of the N-terminal processed region of TWCA1 has not been determined, but it does not appear to encode a polypeptide, in contrast to the C-terminal region of δ-EhCA1. Thus far, attempts to overexpress δ-EhCA1 in E. coli (and Pichia pastoris and Saccharomyces cerevisiae yeast strains) (D. Olszova, personal communication) for purification and antibody production have met with limited success, and other heterologous host systems are being evaluated (e.g., baculovirus and Xenopus laevis expression systems). Considering the fact that heterologous expression of many proteins has not been possible, we are in the process of designing synthetic peptides for the generation of antibodies in order to answer questions regarding its cellular location and metabolic role in E. huxleyi.
The amino acid sequences of the diatom T. weissflogii (TWCA1) and E. huxleyi (δ-EhCA1) are unique and show a common evolutionary history. In addition, they do not share any significant sequence similarity to members of the alpha, beta, and gamma classes of CAs. Analysis of the unprocessed TWCA1 (containing 72 aa upstream of the cleavage site) suggested that it may be a distant homolog of α-CAs (17). Interestingly, all mammalian CAs belong to the alpha class, and three α-CAs have been identified in the green alga Chlamydomonas reinhardtii. CAs can also be differentiated based upon their cellular localization (cytosolic, membrane associated, or secretory). For example, the salt-resistant p60 protein from the unicellular green alga Dunaliella salina is a structurally novel plasma membrane CA containing duplicated regions homologous to animal and C. reinhardtii CAs but modified with acidic residues conferring tolerance to high salinities (11). These kinds of examples illustrate that the evolutionary and structural diversity of CAs continues to increase and support the need to characterize and localize these isozymes in marine phytoplankton.
Due to our inability to identify any hits for δ-EhCA1 in GenBank, we analyzed the upstream region of δ-EhCA1 using PROPSEARCH. One advantage of PROPSEARCH is that it is somewhat efficient at detecting proteins that may be remote homologs that have very little alignment homology. The results of this search yielded significant similarities with members of the cation diffusion protein family (24), including a very high reliability score (99.6%) for the heavy metal ion transporter CzcD. In Ralstonia sp. strain CH34, the heavy metal ion transporter CzcD is involved in the regulation of the Czc system, the latter of which mediates resistance to zinc, cobalt, and cadmium (5). This protein is a member of the cation diffusion protein family (5), some members of which may play a role in importing trace elements (zinc, cobalt, and cadmium) under certain conditions and/or in the binding and exporting of heavy metal cations when the intracellular concentration becomes too high. This is intriguing, given that CAs are metalloenzymes, and CA enzymes employing each of these metals as cofactors have been described previously (15-17). Considering the unusual structural properties of δ-EhCA1, it is not unreasonable to speculate that this protein has a transporter function related to carbon acquisition and/or ion transport processes. In mammalian cells, one carbonic anhydrase isozyme (CAII) interacts with a chloride-HCO3− antiporter, converting uncharged inorganic carbon (CO2) to a charged form (HCO3−) (37). Interestingly, similar scenarios have been described for prokaryotic transport systems that involve bifunctional proteins possessing an N-terminal transmembrane region with similarity to various transporters and a C-terminal domain showing similarity to beta class carbonic anhydrases (34). It is thus conceivable that E. huxleyi δ-EhCA1 could function in a similar manner, and therefore, this cDNA clone should prove to be an important tool for elucidating novel details related to carbon acquisition in coccolithophorids.
The second CA identified in E. huxleyi, γ-EhCA2, was determined to belong to the gamma class. Gamma class CAs are evolutionarily the most ancient of the CA classes (43), and members of this class have been found in all three domains of life. The primary sequence of γ-EhCA2 was most closely related to the enzymes from rice (42%) and Arabidopsis (41%), with significant identity to the enzyme from Helicosporidium (36%). Helicosporidia are nonphotosynthetic green algae and are the only known intracellular pathogens containing plastids, other than Apicomplexa, which includes the malarial parasite Plasmodium falciparum (40, 41). The γ-CA sequence from the chrysophyte E. huxleyi should contribute novel information for studying the evolutionary transition of free-living autotrophs to a parasitic heterotroph, as appears to be the case in these nonphotosynthetic algae. Knowledge of the catalytic mechanism of the gamma class enzyme has come primarily from studies of Cam from M. thermophila (1, 2, 42). The enzyme from E. huxleyi shows distinct differences from the well-studied Cam protein with regard to the conservation of key residues shown to be essential for catalysis. For example, three His residues coordinate the metal ligand in Cam, and site-directed mutagenesis of Gln-75 yielded an enzyme deficient in CO2 hydration activity. These four corresponding residues were completely absent from the recombinant yet catalytically active E. huxleyi enzyme. As pointed out previously, the enzyme from rice lacks one of the three His residues (His-122), while the closely related enzyme from Arabidopsis (77% identity) contains all three of these residues. In addition, the iron- and cobalt-substituted Cam enzyme was shown to have higher CO2 hydration rates than the zinc form (42), and cobalt- and cadmium-containing CAs in the diatom T. weissflogii have been described previously (15-17). These observations suggest a diversity of mechanisms in gamma class CAs that remain to be elucidated, and the availability of large amounts of recombinant E. huxleyi protein will allow for a detailed structural and kinetic analysis of this novel enzyme.
Answers to questions regarding the relationships between photosynthesis and calcification; the mechanisms involved in the regulation, acquisition, and incorporation of DIC; and the presence or absence of a CCM in coccolithophorids remain elusive. Model systems for investigating these processes are most well developed in prokaryotes, including cyanobacteria (35, 36, 49), chemoautotrophic prokaryotes, (32, 38), and anoxygenic photosynthetic bacteria (3, 10, 38). An understanding of carbon metabolism and CCMs in these organisms came from the use of molecular tools, the development of genetic manipulation techniques, and the generation of genotypic and/or phenotypic mutants to decipher functions and pathways. With the exception of RubisCO (largely used as a phylogenetic marker), this is the first report to describe the identification and regulation of any genes or proteins involved in carbon metabolism in coccolithophorids. The study of inorganic carbon acquisition and metabolism in coccolithophorid algae is therefore in its infancy with regard to molecular and biochemical methodologies. In this report, we have described the identification and preliminary characterization of two genetically, structurally, and biochemically distinct carbonic anhydrases from E. huxleyi. In addition, we have presented evidence indicating that transcripts of these isozymes are differentially expressed during the cell cycle and growth phases. These data should provide an important tool for resolving the debates and questions on inorganic carbon metabolism and for increasing our understanding of the biology and ecology of these fascinating marine microalgae.
Acknowledgments
This work has been supported by MBRS-SCORE grant GM 05983 from the National Institutes of Health.
We thank Francois Morel for providing diatom cadmium carbonic anhydrase sequence data and TWCA1 antiserum, both of which were helpful in this study. We also thank William Whalen for the design of Fig. 1 and Sandy Sakulterdkiat and Daniela Olszova for their helpful input on the manuscript.
REFERENCES
- 1.Alber, B. E., C. M. Colangelo, J. Dong, C. M. Stalhandske, T. T. Baird, C. Tu, C. A. Fierke, D. N. Silverman, R. A. Scott, and J. G. Ferry. 1999. Kinetic and spectroscopic characterization of the gamma-carbonic anhydrase from the methanoarchaeon Methanosarcina thermophila. Biochemistry 38:13119-13128. [DOI] [PubMed] [Google Scholar]
- 2.Alber, B. E., and J. G. Ferry. 1994. A carbonic anhydrase from archaeon Methanosarcina thermophila. Proc. Natl. Acad. Sci. USA 91:6909-6913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Allgaier, M., H. Uphoff, A. Felske, and I. Wagner-Döbler. 2003. Aerobic anoxygenic photosynthesis in Roseobacter clade bacteria from diverse marine habitats. Appl. Environ. Microbiol. 69:5051-5059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Anning, T., N. Nimer, M. J. Merrett, and C. Brownlee. 1996. Costs and benefits of calcification in coccolithophorids. J. Mar. Syst. 9:45-56. [Google Scholar]
- 5.Anton, A., C. Große, J. Reißmann, T. Pribyl, and D. H. Nies. 1999. CzcD is a heavy metal ion transporter involved in regulation of heavy metal resistance in Ralstonia sp. strain CH34. J. Bacteriol. 181:6876-6881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Badger, M. R., T. J. Andrews, S. M. Whitney, M. Lugwig, D. C. Yellowlees, W. Leggat, and G. D. Price. 1998. The diversity and coevolution of Rubisco, plastids, pyrenoids, and chloroplast-based CO2-concentrating mechanisms in algae. Can. J. Bot. 76:1052-1071. [Google Scholar]
- 7.Berchner-Pfannschmidt, U., F. Petrat, K. Doege, B. Trinidad, P. Freitag, E. Metzen, H. de Groot, and J. Fandrey. 2004. Chelation of cellular calcium modulates hypoxia-inducible gene expression through activation of hypoxia-inducible factor-1α. J. Biol. Chem. 279:44976-44986. [DOI] [PubMed] [Google Scholar]
- 8.Brownlee, C., N. Nimer, L. F. Dong, and M. J. Merrett. 1994. Cellular regulation during calcification in Emiliania huxleyi. Syst. Assoc. Spec. Vol. 51:133-148. [Google Scholar]
- 9.Buitenhuis, E. T., K. R. Timmermans, and J. W. de Baar. 2003. Zinc-bicarbonate colimitation of Emiliania huxleyi. Limnol. Oceanogr. 48:1575-1582. [Google Scholar]
- 10.Eisen, J. A., K. E. Nelson, I. T. Paulsen, J. F. Heidelberg, M. Wu, R. J. Dodson, R. Deboy, M. L. Gwinn, W. C. Nelson, D. H. Haft, E. K. Hickey, J. D. Peterson, A. S. Durkin, J. L. Kolonay, F. Yang, I. Holt, L. A. Umayam, T. Mason, M. Brenner, T. P. Shea, D. Parksey, W. C. Nierman, T. V. Feldblyum, C. L. Hansen, M. B. Craven, D. Radune, J. Vamathevan, H. Khouri, O. White, T. M. Gruber, K. A. Ketchum, J. C. Venter, H. Tettelin, D. A. Bryant, and C. M. Fraser. 2002. The complete genome sequence of. Chlorobium tepidum TLS, a photosynthetic, anaerobic, green-sulfur bacterium. Proc. Natl. Acad. Sci. USA 99:9509-9514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fisher, M., I. Gokhman, U. Pick, and A. Zamir. 1996. A salt-resistant plasma membrane carbonic anhydrase is induced by salt in Dunaliella salina. J. Biol. Chem. 271:17718-17723. [DOI] [PubMed] [Google Scholar]
- 12.Giordano, M., A. Norici, M. Forssen, M. Eriksson, and J. A. Raven. 2003. An anaplerotic role for mitochondrial carbonic anhydrase in Chlamydomonas reinhardtii. Plant Physiol. 132:2126-2134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Große, C., G. Grass, A. Anton, S. Franke, A. N. Santos, B. Lawley, N. L. Brown, and D. H. Nies. 1999. Transcriptional organization of the czc heavy-metal homeostasis determinant from Alcaligenes eutrophus. J. Bacteriol. 181:2385-2393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guillard, R. R. L. 1975. Culture of phytoplankton for feeding marine invertebrates, p. 29-60. In W. L. Smith and M. H. Chanley (ed.), Culture of marine invertebrate animals. Plenum Press, New York, N.Y.
- 15.Lane, T., M. A. Saito, G. N. George, I. J. Pickering, R. C. Prince, and F. F. M. Morel. 2005. Isolation and preliminary characterization of a cadmium carbonic anhydrase from a marine diatom. Nature 435:42. [DOI] [PubMed] [Google Scholar]
- 16.Lane, T. W., and F. M. M. Morel. 2000. A biological function for cadmium in marine diatoms. Proc. Natl. Acad. Sci. USA 97:4627-4631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lane, T. W., and F. M. M. Morel. 2000. Regulation of carbonic anhydrase by zinc, cobalt, and carbon dioxide in the marine diatom Thalassiosira weissflogii. Plant Physiol. 123:345-352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Marcus, E. A., A. P. Moshfegh, G. Sachs, and D. R. Scott. 2005. The periplasmic α-carbonic anhydrase activity of Helicobacter pylori is essential for acid acclimation. J. Bacteriol. 187:729-738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mitra, M., S. M. Lato, R. A. Ynalvez, Y. Xiao, and J. V. Moroney. 2004. Identification of a new chloroplast carbonic anhydrase in Chlamydomonas reinhardtii. Plant Physiol. 135:173-182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nguyen, B., R. M. Bowers, T. M. Wahlund, and B. A. Read. 2005. Suppressive subtractive hybridization of and differences in gene expression content of calcifying and noncalcifying cultures of Emiliania huxleyi strain 1516. Appl. Environ. Microbiol. 71:2564-2575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nimer, N. A., C. Brownlee, and M. J. Merrett. 1994. Carbon dioxide availability, intracellular pH and growth rate of the coccolithophore Emiliania huxleyi. Mar. Ecol. Prog. Ser. 109:257-262. [Google Scholar]
- 22.Nimer, N. A., Q. Guan, and M. J. Merrett. 1994. Extra- and intra-cellular carbonic anhydrase in relation to culture age in a high-calcifying strain of Emiliania huxleyi Lohmann. New Phytol. 126:601-607. [Google Scholar]
- 23.Paasche, E. 2002. A review of the coccolithophorid Emiliania huxleyi (Prymnesiophyceae), with particular reference to growth, coccolith formation, and calcification-photosynthesis interactions. Phycol. Rev. 40:503-529. [Google Scholar]
- 24.Paulsen, I. T., and M. H. Saier, Jr. 1997. A novel family of ubiquitous heavy metal ion transport proteins. J. Membr. Biol. 156:99-103. [DOI] [PubMed] [Google Scholar]
- 24a.Quinn, P., R. M. Bowers, X. Zhang, T. M. Wahlund, M. A. Fanelli, D. Olszova, and B. A. Read. 2006. cDNA microarrays as a tool for identification of biomineralization proteins in the coccolithophorid Emiliania huxleyi (Haptophyta). Appl. Environ. Microbiol. 72:5512-5526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Raven, J. A. 1997. Putting the C in phycology. Eur. J. Phycol. 32:319-333. [Google Scholar]
- 26.Reinfelder, J. R., A. M. Kraepiel, and F. M. Morel. 2000. Unicellular C4 photosynthesis in a marine diatom. Nature 407:996-999. [DOI] [PubMed] [Google Scholar]
- 27.Roberts, S. B., T. W. Lane, and F. M. M. Morel. 1997. Carbonic anhydrase in the marine diatom Thalassiosira weissflogii (Bacillariophyceae). J. Phycol. 33:845-850. [Google Scholar]
- 28.Rost, B., U. Riebesell, and S. Burkardt. 2003. Carbon acquisition in bloom-forming phytoplankton. Limnol. Oceanogr. 48:55-67. [Google Scholar]
- 29.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 30.Satoh, D., Y. Hiraoka, B. Colman, and Y. Matsuda. 2001. Physiological and molecular biological characterization of intracellular carbonic anhydrase from the marine diatom Phaeodactylum tricornutum. Plant Physiol. 126:1459-1470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shiraiwa, Y. 2003. Physiological regulation of carbon fixation in the photosynthesis and calcification of coccolithophorids. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 136:775-783. [DOI] [PubMed] [Google Scholar]
- 32.Shively, J. M., G. van Keulen, and W. G. Meijer. 1998. Something from almost nothing: carbon dioxide fixation in chemoautotrophs. Annu. Rev. Microbiol. 52:191-230. [DOI] [PubMed] [Google Scholar]
- 33.Sikes, C. S., and V. J. Fabry. 1994. Photosynthesis, CaCO3 deposition, coccolithophorids and the global carbon cycle. >In N. E. Tolbert and J. Preiss (ed.), Photosynthetic carbon metabolism and regulation of atmospheric CO2 and O2. Oxford University Press, Oxford, United Kingdom.
- 34.Smith, K. S., and J. G. Ferry. 2000. Prokaryotic carbonic anhydrases. FEMS Microbiol. Rev. 24:335-366. [DOI] [PubMed] [Google Scholar]
- 35.So, A. K.-C., G. S. Espie, E. B. Williams, J. M. Shively, S. Heinhorst, and G. C. Cannon. 2004. A novel evolutionary lineage of carbonic anhydrase (ɛ class) is a component of the carboxysome shell. J. Bacteriol. 186:623-630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stal, L. 1995. Physiological ecology of cyanobacteria in microbial mats and other communities. New Phytol. 131:1-32. [DOI] [PubMed] [Google Scholar]
- 37.Sterling, D., R. A. F. Reithmeier, and J. R. Casey. 2001. A transport metabolon. Functional interaction of carbonic anhydrase II and chloride/bicarbonate exchangers. J. Biol. Chem. 276:47886-47894. [DOI] [PubMed] [Google Scholar]
- 38.Tabita, F. R. 1995. The biochemistry and metabolic regulation of carbon metabolism and CO2 fixation in purple bacteria, p. 885-914. In R. E. Blankenship, M. T. Madigan, and C. E. Bauer (ed.), Anoxygenic photosynthetic bacteria. Kluwer Academic Publishers, Dordrecht, The Netherlands.
- 39.Tanaka, Y., D. Nakatsuma, H. Harada, M. Ishida, and Y. Matsuda. 2005. Localization of soluble β-carbonic anhydrase in the marine diatom Phaeodactylum tricornutum. Sorting to the chloroplast and cluster formation on the girdle lamellae. Plant Physiol. 138:207-217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tartar, A., and D. G. Boucias. 2004. The non-photosynthetic, pathogenic green alga Helicosporidium sp. has retained a modified, functional plastid genome. FEMS Microbiol. Lett. 233:153-157. [DOI] [PubMed] [Google Scholar]
- 41.Tartar, A., D. G. Boucias, B. J. Adams, and J. J. Becnel. 2002. Phylogenetic analysis identifies the invertebrate pathogen Helicosporidium sp. as a green alga (Chlorophyta). Int. J. Syst. Evol. Microbiol. 52:273-279. [DOI] [PubMed] [Google Scholar]
- 42.Tripp, B. C., C. B. Bell III, F. Cruz, C. Krebs, and J. G. Ferry. 2004. A role for iron in an ancient carbonic anhydrase. J. Biol. Chem. 279:6683-6687. [DOI] [PubMed] [Google Scholar]
- 43.Tripp, B. C., K. Smith, and J. G. Ferry. 2001. Carbonic anhydrase: new insights for an ancient enzyme. J. Biol. Chem. 276:48615-48618. [DOI] [PubMed] [Google Scholar]
- 44.Voznesenskaya, E. V., V. R. Franceschi, O. Kiirats, H. Freitag, and G. E. Edwards. 2001. Kranz anatomy is not essential for terrestrial C4 plant photosynthesis. Nature 414:543-546. [DOI] [PubMed] [Google Scholar]
- 45.Wahlund, T. M., A. R. Hadaegh, R. Clark, B. Nguyen, M. Fanelli, and B. A. Read. 2004. Analysis of expressed sequence tags from calcifying cells of the marine coccolithophorid, Emiliania huxleyi. Mar. Biotechnol. 6:278-290. [DOI] [PubMed] [Google Scholar]
- 46.Wahlund, T. M., X. Zhang, and B. A. Read. 2005. Expressed sequence tag profiles from calcifying and non-calcifying cultures of Emiliania huxleyi. J. Micropaleontol. 50:145-155. [Google Scholar]
- 47.Wawrik, B., J. H. Paul, and F. R. Tabita. 2002. Real-time PCR quantification of rbcL (ribulose-1,5-bisphosphate carboxylase/oxygenase) mRNA in diatoms and pelagophytes. Appl. Environ. Microbiol. 68:3771-3779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wilbur, K. M., and N. G. Anderson. 1948. Electrometric and colorimetric determination of carbonic anhydrase. J. Biol. Chem. 176:147-154. [PubMed] [Google Scholar]
- 49.Woodger, F. J., M. R. Badger, and G. D. Price. 2003. Inorganic carbon limitation induces transcripts encoding components of the CO2-concentrating mechanism in Synechococcus sp. PCC7942 through a redox-independent pathway. Plant Physiol. 133:2069-2080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wyman, M., J. T. Davies, D. W. Crawford, and D. A. Purdie. 2000. Molecular and physiological responses of two classes of marine chromophytic phytoplankton (diatoms and prymnesiophytes) during the development of nutrient-stimulated blooms. Appl. Environ. Microbiol. 66:2349-2357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wyman, M., J. T. Davies, S. Hodgson, G. A. Tarran, and D. A. Purdie. 2005. Dynamics of ribulose 1,5-bisphosphate carboxylase/oxygenase gene expression in the coccolithophorid Coccolithus pelagicus during a tracer release experiment in the Northeast Atlantic. Appl. Environ. Microbiol. 71:1659-1661. [DOI] [PMC free article] [PubMed] [Google Scholar]