Abstract
Flooded rice fields have become a model system for the study of soil microbial ecology. In Italian rice fields, in particular, aspects from biogeochemistry to molecular ecology have been studied, but the impact of protistan grazing on the structure and function of the prokaryotic community has not been examined yet. We compared an untreated control soil with a γ-radiation-sterilized soil that had been reinoculated with a natural bacterial assemblage. In order to verify that the observed effects were due to protistan grazing and did not result from sterilization, we set up a third set of microcosms containing sterilized soil that had been reinoculated with natural assemblage bacteria plus protists. The spatial and temporal changes in the protistan and prokaryotic communities were examined by denaturing gradient gel electrophoresis (DGGE) and terminal restriction fragment length polymorphism (T-RFLP) analysis, respectively, both based on the small-subunit gene. Sequences retrieved from DGGE bands were preferentially affiliated with Cercozoa and other bacteriovorous flagellates. Without protists, the level of total DNA increased with incubation time, indicating that the level of the microbial biomass was elevated. Betaproteobacteria were preferentially preyed upon, while low-G+C-content gram-positive bacteria became more dominant under grazing pressure. The bacterial diversity detectable by T-RFLP analysis was greater in the presence of protists. The level of extractable NH4+ was lower and the level of extractable SO42− was higher without protists, indicating that nitrogen mineralization and SO42− reduction were stimulated by protists. Most of these effects were more obvious in the partially oxic surface layer (0 to 3 mm), but they could also be detected in the anoxic subsurface layer (10 to 13 mm). Our observations fit well into the overall framework developed for protistan grazing, but with some modifications pertinent to the wetland situation: O2 was a major control, and O2 availability may have limited directly and indirectly the development of protists. Although detectable in the lower anoxic layer, grazing effects were much more obvious in the partially oxic surface layer.
Wetland soils are characterized by unique biogeochemical cycles and by unique microbial communities compared to upland soils (16, 38, 49). The most important controls for microbial activity are organic matter input and the restricted availability of O2. Both of these factors depend on the dominant vegetation, which may supply the soil not only with organic matter but also to a significant but varying extent with O2 (28, 34). The same controls act on natural and man-made wetlands, and among the latter rice fields are by far the best-studied ecosystems (16, 37). Significant progress has been made in understanding the interaction between rice plants, soil biogeochemistry, and microbes (7, 8, 16, 37, 43), and thus rice fields are one of the best-studied model systems in soil microbial ecology. However, most work published so far has ignored the role of microbial mortality. From recent work on ciliates it became evident that grazing may have an effect on soil bacteria in rice fields (57, 58). However, ciliates were outnumbered by flagellates, which might have a much greater effect on microbes (58).
Much more is known about the effects of protists on planktonic bacteria. Under grazing pressure, bacteria may change their growth form (32, 36), and the community composition may also change (52). Moreover, protists may control microbial net production and the proportion of active bacteria (19). Significant work has also been done with upland soils; grazing has been shown to accelerate nutrient cycling and to have far-reaching effects even at the plant community level (9, 15).
All this evidence let us expect similar effects in flooded rice field soils. Hence, we examined the effects of protists on the structure and activity of the bacterial community in a water-saturated rice soil. We used soil from a rice field in Vercelli (Italy) in which the diversity and dynamics of different groups of bacteria are already known (23, 29, 40, 56, 60). In particular, the succession of bacteria across the oxygen gradient at the soil surface has been studied in great detail (51).
A common technique for studying grazing effects is the use of metabolic inhibitors. However, the effective use of inhibitors in a complex community requires that they be specific to their target group without affecting the nontarget organisms (55). For eukaryotes, the inhibitors that are most frequently used are cycloheximide and colchicine. However, the specificity of cycloheximide has been found to be inadequate (55, 59), and colchicine may inhibit methanogenesis (12). Furthermore, cells killed by the inhibitors may serve as substrates for surviving microorganisms, and the inhibitors may be degraded during prolonged incubation (4). Therefore, we compared an untreated control soil with a γ-radiation-sterilized soil that had been reinoculated with a natural bacterial assemblage. In order to verify that the observed effects were due to protistan grazing and did not result from sterilization, we set up a third set of microcosms containing sterilized soil that had been reinoculated with bacteria plus protists. In order to determine the effect of oxygen gradients on both the microbial (51) and protistan communities, we analyzed two layers, a partially oxic upper layer (0 to 3 mm) and a totally anoxic lower layer (10 to 13 mm). Eukaryotic and bacterial communities were analyzed by molecular methods. Respiration, carbon and nitrogen mineralization, and porewater chemistry were used to study the gross effect of protists on biogeochemical functions.
MATERIALS AND METHODS
Soil and field site.
Soil was taken from a rice field of the Istituto Sperimentale della Risicoltura (Vercelli, Italy) in spring 2000 before flooding. The field site and soil properties have described previously (33, 39). The soil was air dried and stored as dry lumps at room temperature. Prior to use, the soil was ground with a jaw crusher (Retsch, Hahn, Germany) to obtain particles that were <2 mm in diameter.
Microcosms and inocula.
Soil was sterilized by γ irradiation (25 kGy; 60Co; Zentrale Strahlenschutzgruppe der Justus-Liebig-Universität, Giessen, Germany). When slurried and incubated anaerobically for 2 weeks, γ-irradiated soil showed neither CO2 production nor CH4 production. Microcosms were prepared by reinoculating the sterilized soil with indigenous protistan and bacterial assemblages prepared as described below. Twenty grams of the sterilized soil was added to a serum bottle (inside diameter, 3.4 cm; volume, 60 cm3), forming a soil layer 15 mm deep. The soil was inoculated with protists and bacteria (treatment P+B) or with only bacteria (treatment B) while the headspace was flushed with N2 to minimize the detrimental effect of O2 on the anaerobic microorganisms. Aerobic microorganisms were assumed to tolerate the anoxia during preparation of inocula and microcosms, which was completed in ≤10 h. For comparison, unsterilized soil was inoculated with a filter-sterilized inoculum and served as a control. The inoculum (8 ml) water logged the soil completely. However, the soil was not flooded to exclude protists that might have grown in the overlying water. The bottles were plugged with butyl rubber stoppers and aluminum caps. Bubbles entrapped in the soil were removed, and the soil surface was flattened by gently knocking the bottom of the bottle on a table. Altogether, 45 bottles were prepared. The headspace was flushed using hypodermic needles with water-saturated and filter-sterilized air for 5 min, and the bottles were incubated at 25°C in the dark. Flushing was done at the beginning and after 2, 6, 13, and 20 days.
For preparation of protistan and bacterial inocula, 2 kg of soil was mixed with 1 liter of distilled water and incubated for 30 days at 25°C in the dark. Autoclaved O2-free water was used to extract microorganisms from the soil by using the following procedures. The top 15 mm of the incubated soil was mixed with water at ratios of 1:1 (wt/wt) (protists) and 1:2 (bacteria). The soil slurry for preparation of the protistan inoculum was shaken manually and allowed to settle for 15 min before the supernatant was collected (treatment S) (Table 1). The soil slurry used for preparation of the bacterial mixture was homogenized with a Waring blender for 1 min. In a pilot experiment this treatment gave the best recovery of bacteria (data not shown). The supernatant was obtained after centrifugation at 1,500 × g for 10 min (treatment C) (Table 1). The supernatants were subjected to the following successive filtration steps: (i) a 200-μm mesh sieve to exclude coarse soil particles, which most protists and detached bacteria could pass through; (ii) a 3-μm Nuclepore filter to exclude protists; and (iii) a 0.2-μm membrane filter to exclude protists and bacteria. Finally, three different inocula were prepared from the appropriate size fractions, as summarized in Table 1. The mixing ratio of the preparations obtained from treatments S and C was 1:1 (vol/vol). All steps were carried out with N2 flushing, and the inocula were stored at 4°C under N2 until use.
TABLE 1.
Preparation of inoculaa
| Soil | Inoculum for treatment | Previous treatmentb | Fractions
|
||
|---|---|---|---|---|---|
| <200 μm | <3 μm | <0.2 μm | |||
| γ-Irradiated | P+B | S | + | − | − |
| C | − | + | − | ||
| γ-Irradiated | B | S | − | + | − |
| C | − | + | − | ||
| Untreated | Control | S | − | − | + |
| C | − | − | + | ||
For further details see the text.
S, suspension and settling of the supernatant; C, blending and centrifugation of the supernatant.
Biogeochemistry.
Gas samples (50 μl) were taken before and 2 to 3 days after the headspace was flushed. CO2 was measured with a gas chromatograph equipped with a methanizer and a flame ionization detector after conversion to methane, and O2 was measured with a gas chromatograph equipped with a thermal conductivity detector. The rates of CO2 accumulation and O2 consumption were calculated from the changes in gas concentrations.
O2 microprofiles were measured with a Clark-type microelectrode (OX 100 with PA2000 picoammeter; Unisense AS, Denmark) the day after the headspace was flushed. The butyl rubber stopper and aluminum seal were removed just before measurement. The electrodes were mounted on a micromanipulator with a vertical resolution of 10 μm. A dissecting microscope was used to adjust the tip of the microelectrode with respect to the soil surface. The O2 profiles were determined with a vertical resolution of 100 μm down to a depth of 1,500 μm.
Triplicate bottles which received each treatment were sacrificed after the O2 profiles had been determined. The bottles were shock frozen in liquid N2 and broken. The frozen soil was sectioned into 200-μm slices using a precooled (−20°C) microtome (Microm, Walldorf, Germany). From each bottle, slices from the top layer (0 to 3 mm, termed the upper layer) and from a deeper layer (10 to 13 mm, termed the lower layer) were pooled and stored at −20°C until further analysis. Chemical and molecular analyses were performed with aliquots of the same soil.
Exchangeable ammonium was extracted from 0.15 g (wet weight) of soil after it was mixed with 1.5 ml of 2 N KCl. The extract was centrifuged (14,000 × g, 5 min) and filtered (<0.2 μm). The NH4+ concentration was determined fluorometrically (26) by microscale analysis. The extract was mixed with 0.25 volume of a reaction buffer (15 mM o-phthalaldehyde and 50 mM 2-mercaptoethanol in 500 mM purified phosphate buffer, pH 6.8) (17) in 96-well microtiter plates and incubated at 63°C for 10 min. After cooling to room temperature, the fluorescence intensity was determined at an excitation wavelength of 410 nm and an emission wavelength of 470 nm with a SAFIRE microplate reader (TECAN, Crailsheim, Germany). Nitrate and sulfate in the soil were extracted from 0.15 g (wet weight) of soil by mixing the soil with 600 μl of distilled water for 1 h. The extract was obtained as described above, and the nitrate and sulfate contents were determined by ion chromatography (5).
Molecular analyses.
DNA was extracted from 0.3 g (wet weight) of soil with a Fast DNA SPIN kit (Bio 101, La Jolla, CA) used according to the manufacturer's instructions but with an additional washing step with guanidine isothiocyanate. DNA was eluted from the binding matrix with 100 μl of DNase-free water and stored at −20°C. DNA concentrations were determined fluorometrically using a PicoGreen double-stranded DNA (dsDNA) quantitation kit (Molecular Probes, Leiden, The Netherlands) in 96-well microtiter plates with a microplate reader.
The eukaryotic community was analyzed by denaturing gradient gel electrophoresis (DGGE) as described by Díez et al. (20). A fragment of the 18S rRNA gene (approximately 560 bp) was amplified from 2 μl of environmental DNA with primers Euk1A and Euk516r-GC (20). The reaction mixture (100 μl) contained 50 pmol of each primer, each deoxyribonucleoside triphosphate at a concentration of 200 μM, 400 ng μl−1 of bovine serum albumin (Roche Diagonistics, Mannheim, Germany), 1.5 mM MgCl2 (Promega, Madison, WI), 2.5 U Taq polymerase (Promega), and 0.1 volume of a 10× PCR buffer provided with the enzyme. The PCR program included an initial denaturation step of 130 s at 94°C, followed by 25 cycles of denaturation (30 s, 94°C), primer annealing (45 s, 56°C), and primer extension (130 s, 72°C) and a final extension step of 7 min at 72°C with a thermal cycler (Eppendorf Mastercycler gradient; Eppendorf, Hamburg, Germany).
DGGE of the amplified 18S rRNA gene fragments was performed with a DCode system (Bio-Rad, California) by using 1-mm-thick 6% polyacrylamide gels and a 20 to 50% denaturant gradient (100% denaturant contained 7 M urea and 40% [vol/vol] formamide). Ten microliters of PCR products was loaded onto the gels, and electrophoresis was carried out in 1× Tris-acetate-EDTA buffer at 100 V for 16 h at a constant temperature (60°C). The gels were stained with 1:10,000 (vol/vol) SYBR Green I (Biozym, Hessisch-Oldendorf, Germany) for 30 min and scanned with a Storm 860 phosphorimager (Molecular Dynamics, Sunnyvale, CA).
DGGE bands were excised from the DGGE gel and reamplified, the correct mobility on a DGGE gel was verified, and the bands were sequenced. Sequencing reactions were performed with an ABI PRISM BigDye terminator version 1.1 cycle sequencing kit (Applied Biosystems, California) used according to the manufacturer's instruction, using the same primer set but without the GC clamp. The cycle sequencing products were analyzed with an ABI 377 DNA sequencer. Sequences were compared to the NCBI database using BlastN searches.
The bacterial community was analyzed by the terminal restriction fragment length polymorphism (T-RFLP) method. The 16S rRNA genes were amplified from 0.1 μl of the environmental DNA extract using primers 8-27F (21) and 1392-1407R (42). The forward primer was 5′ labeled with 6-carboxyfluorescein. The reaction mixture (50 μl) contained 10 pmol of each primer, each deoxyribonucleoside triphosphate at a concentration of 200 μM, 400 ng of bovine serum albumin (Roche Diagonistics) μl−1, 1.5 mM MgCl2 (Promega), 0.5 U of Taq polymerase (Promega), and 0.1 volume of a 10× PCR buffer provided with the enzyme. The PCR program included an initial denaturation step of 5 min at 95°C, followed by 30 cycles of denaturation (1 min, 95°C), primer annealing (1 min, 57°C), and primer extension (3 min, 72°C) and a final extension step of 7 min at 72°C. The PCR products were purified with a QIAquick PCR purification kit (QIAGEN, Hilden, Germany). The concentration of the purified products was determined photometrically.
Aliquots of the amplicons (50 ng) were digested with 3 U of MspI (C'CGG; Promega) for 2 h at 37°C in a 10-μl (total volume) reaction mixture containing 1 μl of the 10× incubation buffer (Promega) and 1 μl of bovine serum albumin. Fluorescently labeled terminal restriction fragments (T-RFs) were size separated with an ABI 373A automated sequencer (PE Applied Biosystems). T-RFLP electropherograms were analyzed by determining peak height (GeneScan 2.1 software; PE Applied Biosystems). The percent abundance (Ap) of a T-RF was calculated by comparison with the total fluorescence intensity of all T-RFs in the sample. Only T-RFs that were between 50 and 900 bp long and had an Ap of >1% in any sample were included in further calculations. The phylogenetic affiliation of the T-RFs was based on a library containing 190 clones of the eubacterial 16S rRNA and its gene obtained previously from the same soil (51).
Statistical analyses.
Differences between treatments or time of incubation were tested by using a one-way analysis of variance (SPSS for Windows, version 10.0). The T-RFLP profiles were compared by correspondence analysis using CANOCO (version 4.5; Microcomputer Power, Inc., Ithaca, NY).
Nucleotide sequence accession numbers.
The 18S rRNA gene sequences of the DGGE bands have been deposited in the DDBJ, EMBL, and GenBank nucleotide sequence databases under accession no. AB222279 to AB222351.
RESULTS
Biogeochemistry.
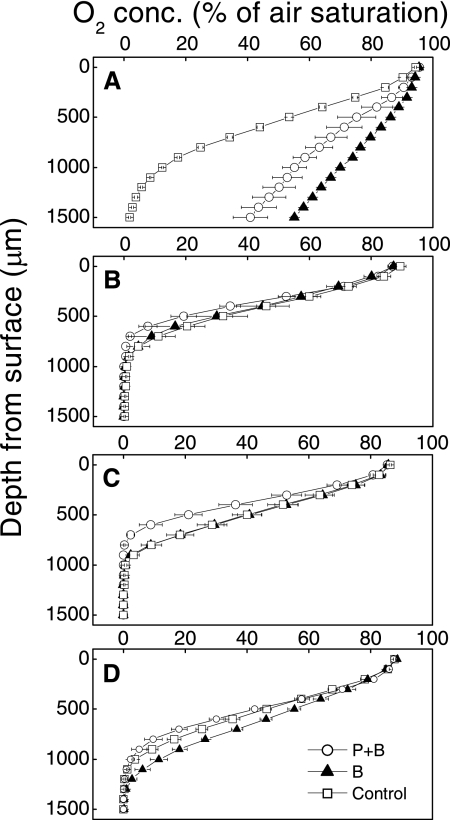
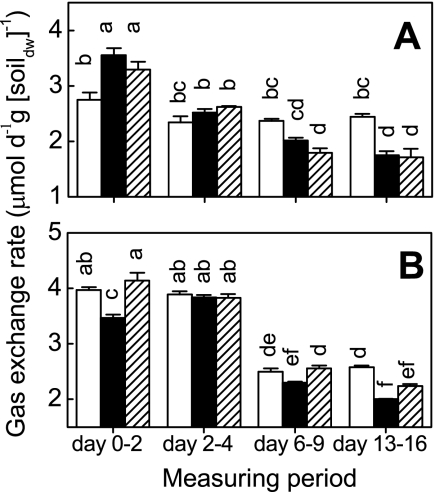
As soon as 10 h after flooding, the porewater O2 saturation in the control microcosms dropped to nearly zero at a depth of 1.5 mm (Fig. 1). In the reinoculated microcosms (treatments B and P+B), however, the O2 saturation at the same depth was >40%, corresponding to >100 μM (Fig. 1A). During the first 2 days, the treatment B microcosms respired as much O2 as the control, while the levels of respiration in the treatment P+B microcosms were significantly lower (Fig. 2A). After 7 days the oxic zone was not deeper than 0.9 mm, and the profiles were very similar for all microcosms, with the standard errors overlapping (Fig. 1B). O2 uptake remained constant with time for treatment P+B but decreased for the other treatments (Fig. 2A). CO2 emission was highest during the first 2 days and decreased later to about 50% (Fig. 2B). In addition to this common trend, the treatment B microcosms emitted less CO2.
FIG. 1.
Effects of different inocula on O2 gradients measured after 10 h (A), 7 days (B), 14 days (C), and 21 days (D). The values are means ± standard errors (n = 3).
FIG. 2.
Effects of different microbial inocula on O2 consumption (A) and CO2 emission (B). Open bars, sterilized soil inoculated with protists and bacteria (treatment P+B); solid bars, sterilized soil inoculated with bacteria (treatment B); cross-hatched bars, control. The values are means ± standard errors (n = 3). Columns marked with different letters are significantly different (P < 0.05, as determined by Tukey's honestly significant difference test). dw, dry weight.
The level of exchangeable NH4+ was low (0.6 to 1.1 μmol · g [dry weight] of soil−1) at day 0, but it increased with time (Fig. 3). The NH4+ levels in the upper and lower layers were not significantly different within the treatments. The levels of NH4+ were not different for different treatments at day 0, but at days 7, 14, and 21 soil from treatment B microcosms contained significantly less exchangeable NH4+ than soil from the control and treatment P+B microcosms contained (Fig. 3).
FIG. 3.
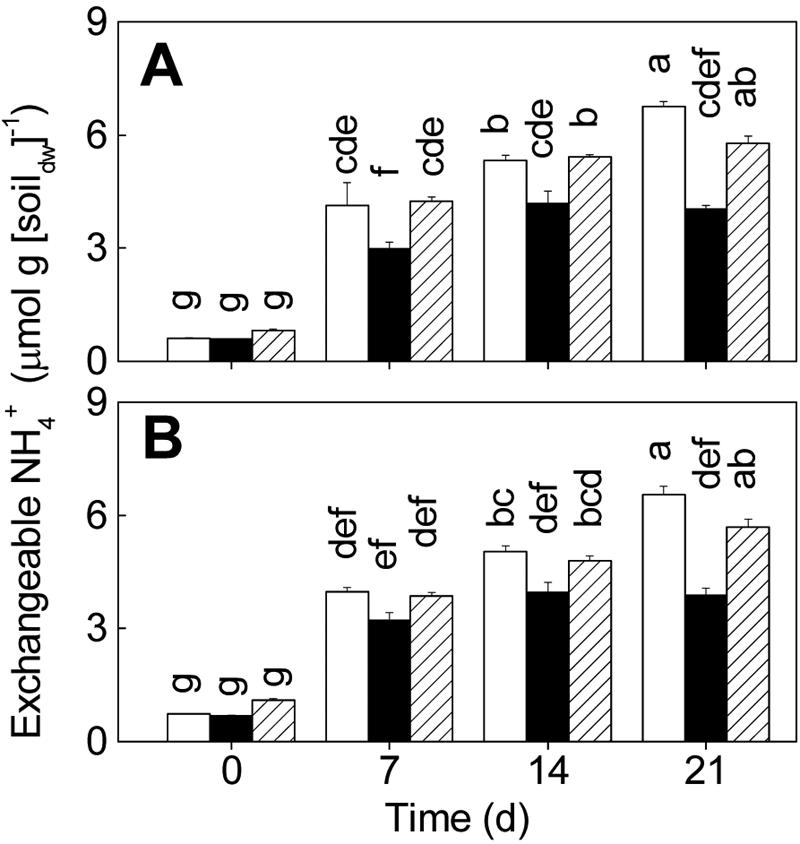
Effects of different microbial inocula on extractable NH4+ levels in the upper layer (0 to 3 mm) (A) and the lower layer (10 to 13 mm) (B). Open bars, sterilized soil inoculated with protists and bacteria (treatment P+B); solid bars, sterilized soil inoculated with bacteria (treatment B); cross-hatched bars, control. The values are means ± standard errors (n = 3). Columns marked with different letters are significantly different (P < 0.05, as determined by Tukey's honestly significant difference test). dw, dry weight.
Soil from the control microcosms contained <0.01 μmol NO3− · g (dry weight) of soil−1 at day 0 (detection limit), while soil from the treatment B and P+B microcosms contained 0.7 to 1.1 μmol NO3− · g (dry weight) of soil−1 (data not shown). From day 7 onward, the NO3− contents in soil from the control microcosms and microcosms that received both treatments were <0.01 μmol · g (dry weight) of soil−1. The amount of extractable SO42− decreased with time in both the upper and lower layers (Fig. 4). The soil from treatment B microcosms contained significantly more SO42− than the soil from the microcosms that received the other treatments (14 and 21 days).
FIG. 4.
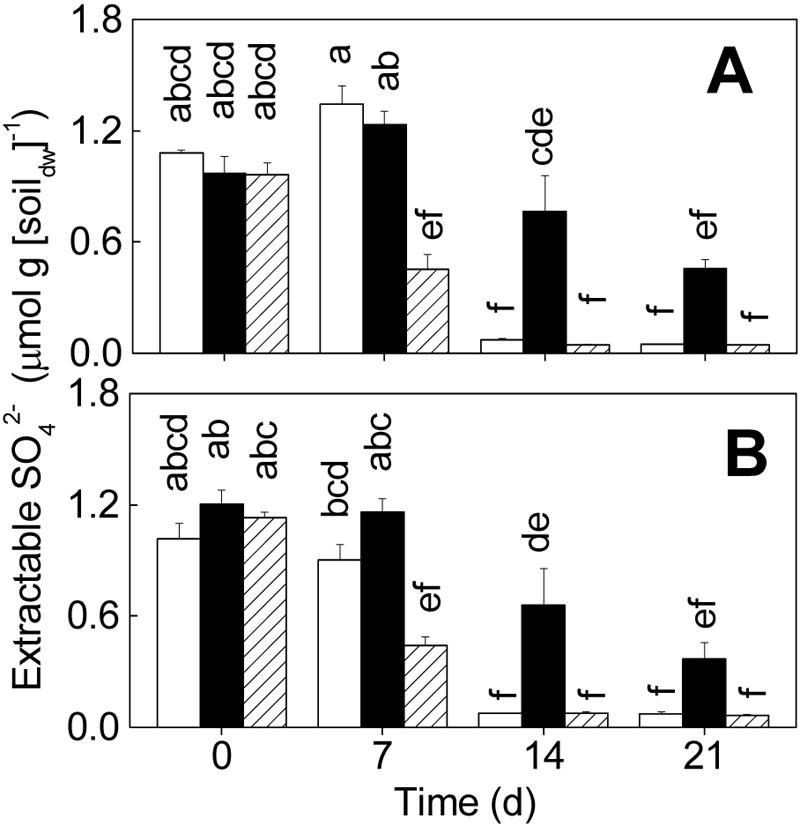
Effects of different microbial inocula on extractable SO42− levels in the upper layer (0 to 3 mm) (A) and the lower layer (10 to 13 mm) (B). Open bars, sterilized soil inoculated with protists and bacteria (treatment P+B); solid bars, sterilized soil inoculated with bacteria (treatment B); cross-hatched bars, control. The values are means ± standard errors (n = 3). Columns marked with different letters are significantly different (P < 0.05, as determined by Tukey's honestly significant difference test). dw, dry weight.
Molecular ecology.
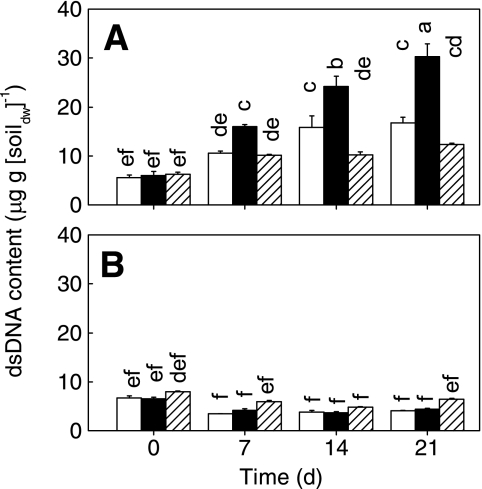
The initial amount of DNA ranged from 5.6 to 8.0 μg · g (dry weight) of soil−1 (Fig. 5). In the lower soil layer, the value stayed nearly constant with time. However, the amount of DNA in the upper layer increased to 10 to 16 μg · g (dry weight) of soil−1 in the microcosms with protists (treatment P+B and control) and to up to 30 μg · g (dry weight) of soil−1 in the treatment B microcosms.
FIG. 5.
Effects of different microbial inocula on the amount of DNA extracted from the upper layer (0 to 3 mm) (A) and the lower layer (10 to 13 mm) (B). Open bars, sterilized soil inoculated with protists and bacteria (treatment P+B); solid bars, sterilized soil inoculated with bacteria (treatment B); cross-hatched bars, control. The values are means ± standard errors (n = 3). Columns marked with different letters are significantly different (P < 0.05, as determined by Tukey's honestly significant difference test). dw, dry weight.
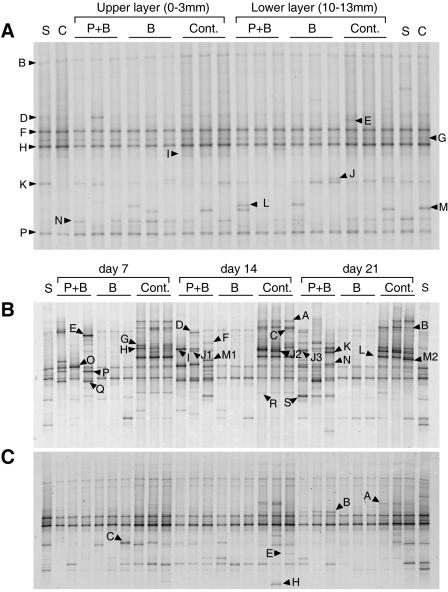
At day 0, the DGGE banding patterns of the 18S rRNA gene amplicons were very similar for all layers and treatments and were also similar to the patterns for the air-dried and sterilized soils (Fig. 6A). All lanes contained bands B, F, G, H, and P (Fig. 6A). No time- or treatment-specific pattern was observed for the other bands. In the upper layer, the number of DGGE bands remained low for treatment B, but the number increased with time for treatment P+B and the control (Fig. 6B). The following three groups of bands could be identified, although some were not observed for all three replicates: (i) bands that cooccurred in treatment P+B and the control (i.e., in all microcosms with protists), including bands D, F, H, J2, K, P, and Q; (ii) bands specific to treatment P+B, including bands E, I, J1, J3, M1, N, O, and S; and (iii) bands specific to the control, including bands A, B, C, G, L, M2, and R. The banding pattern for the lower layer was less diverse, but one band was observed for both treatment P+B and the control (band B on days 14 and 21), and a few bands occurred in only the control (bands A, E, and H) (Fig. 6C).
FIG. 6.
DGGE banding patterns of 18S rRNA gene partial sequences. (A) Comparison of all treatments and layers at day 0. (B and C) Comparison of days 7 to 21 for the upper layer (0 to 3 mm) (B) and the lower layer (10 to 13 mm) (C). Three replicate microcosms were analyzed per day and treatment. P+B, sterilized soil inoculated with protists and bacteria; B, sterilized soil inoculated with bacteria; Cont., control; S, γ-irradiation-sterilized soil; C, intact soil before sterilization. Bands with different mobilities are indicated by different designations. For the phylogenetic affiliation see Table 2. Note that the same designation may indicate different bands in the different gels. Only bands that appeared after day 0 are indicated in panels B and C.
Table 2 summarizes the tentative phylogenetic affiliations of the sequences retrieved from different DGGE bands. Of 40 different sequences, 13 could be affiliated with flagellates, 7 could be affiliated with fungi, 5 could be affiliated with metazoans (including microcrustaceans), 5 could be affiliated with green algae, 4 could be affiliated with higher plants, 3 could be affiliated with ciliates, and 2 could be affiliated with amoebae. Cercozoa was the dominant taxon among the flagellates (11 of 13 sequences). Sixteen of 22 sequences from the upper layer could be assigned to protists. In the lower layer, the nearest match for the sequence retrieved from band B (obtained from treatment P+B and the control) (Fig. 6C) was the match with an anaerobic Cercomonas sp. strain (ATCC 50367).
TABLE 2.
Similarities of sequences obtained from the excised DGGE bands to sequences in the NCBI database
| Banda | Presence in treatment microcosms
|
No. of lanes sequencedb | Closest relative | Accession no.c | % Similarityd | Phylogenetic group | ||
|---|---|---|---|---|---|---|---|---|
| P+B | B | Control | ||||||
| Day 0 | ||||||||
| B | + | + | + | 3 | Spongomonas minima | AF411280 | 99.1 | Flagellates, Cercozoa |
| D | + | + | + | 1 | Ossicaulis lignatilis | AF334923 | 99.4 | Fungi, Basidiomycota |
| E | − | − | + | 1 | Xenillus tegeocranus | AF022042 | 89.3 | Metazoa, Arthropoda |
| F | + | + | + | 2 | Scenedesmaceae sp. | AY197639 | 99.8 | Green algae, Chlorophyta |
| G | + | + | + | 2 | Coelastrella saipanensis | AB055800 | 99.8 | Green algae, Chlorophyta |
| H | + | + | + | 2 | Hydrodictyon reticulatum | M74497 | 100.0 | Green algae, Chlorophyta |
| I | + | + | + | 2 | Hydrodictyon reticulatum | M74497 | 100.0 | Green algae, Chlorophyta |
| J | + | + | 1 | Scrophularia californica | AJ236031 | 98.3 | Plants, Streptophyta | |
| K | + | + | + | 2 | Gilia capitata | L49282 | 99.1-99.2 | Plants, Streptophyta |
| L | + | + | − | 2 | Triticum aestivum | AJ272181 | 99.8 | Plants, Streptophyta |
| M | + | + | + | 2 | Heterocypris vandouwei | AF220460 | 99.3 | Metazoa, Arthropoda |
| N | + | + | + | 2 | Dolerocypris sinensis | AF220459 | 94.5 | Metazoa, Arthropoda |
| P | + | + | + | 2 | Pithophora sp. | AB062713 | 100.0 | Green algae, Chlorophyta |
| Upper layer | ||||||||
| A | − | − | + | 3 | Spathidium sp. | Z22931 | 97.9 | Ciliates, Litostomatea |
| B | − | − | + | 2 | Pseudoplatyophrya nana | AF060452 | 94.9-95.1 | Ciliates, Colpodea |
| C | − | − | + | 3 | Cercomonas sp. | AF534712 | 94.3 | Flagellates, Cercozoa |
| D | + | − | + | 2 | Spongomonas minima | AF411280 | 95.9-96.2 | Flagellates, Cercozoa |
| E | + | − | − | 1 | Cercomonas sp. strain LargeSA | AF411266 | 98.3 | Flagellates, Cercozoa |
| F | + | − | + | 2 | Costa Rica flagellate | AF411277 | 98.5 | Flagellates, Cercozoa |
| G | − | − | + | 2 | Cephaliophora irregularis | AB001109 | 100.0 | Fungi, Ascomycota |
| H | + | − | + | 3 | Powellomyces variabilis | AF164241 | 85.9 | Fungi, Chytridiomycota |
| I | + | − | − | 1 | Cercomonas sp. strain SmallSA | AF534712 | 93.3 | Flagellates, Cercozoa |
| J1 | + | − | − | 1 | Hyphozyma variabilis | AJ496241 | 100.0 | Fungi, Ascomycota |
| J2 | + | − | + | 1 | Boudiera acanthospora | U53373 | 98.4-99.0 | Fungi, Ascomycota |
| J3 | + | − | − | 2 | Bodomorpha minima | AF411276 | 93.4 | Flagellates, Cercozoa |
| K | + | − | + | 1 | Allantion sp. | AF411265 | 89.0 | Flagellates, Cercozoa |
| L | − | − | + | 3 | Ichthyophonus hoferi | AF467801 | 84.2-84.3 | Mesomycetozoa, Ichthyosporea |
| M1 | + | − | − | 3 | Adriamonas peritocrescens | AF243501 | 95.2-96.2 | Flagellates, stramenopiles |
| M2 | − | − | + | 1 | Boudiera acanthospora | U53373 | 98.8 | Fungi, Ascomycota |
| N | + | − | − | 1 | Allantion sp. | AF411265 | 93.9 | Flagellates, Cercozoa |
| O | + | − | − | 1 | Allantion sp. | AF411265 | 91.1 | Flagellates, Cercozoa |
| P | + | − | + | 2 | Echinamoeba exundans | AF293895 | 97.6 | Amoebae, Lobosea |
| Q | + | − | + | 2 | Adriamonas peritocrescens | AF243501 | 94.8 | Flagellates, Stramenopiles |
| R | − | − | + | 2 | Hartmannella cantabrigiensis | AY294147 | 91.4 | Amoebae, Lobosea |
| S | + | − | − | 2 | Sorogena stoianovitchae | AF300287 | 79.4 | Ciliates, Colpodea |
| Lower layer | ||||||||
| A | − | − | + | 1 | Boudiera acanthospora | U53373 | 98.4 | Fungi, Ascomycota |
| B | + | − | + | 4 | Cercomonas sp. | AF411272 | 97.0 | Flagellates, Cercozoa |
| C | − | + | − | 1 | Rhynchospora nervosa | AF207009 | 98.9 | Plants, Streptophyta |
| E | − | − | + | 1 | Heterocypris vandouwei | AF220460 | 96.3 | Metazoa, Arthropoda |
| H | − | − | + | 1 | Leptestheria compleximanus | AF144213 | 100.0 | Metazoa, Arthropoda |
See Figure 6 for the designations of DGGE bands.
Number of lanes from which the same band was sequenced. Note that for the sake of clarity the band is labeled in Fig. 6 in only one lane.
Accession number of the closest relative.
A range is given if the sequences retrieved from different lanes were very similar but not identical.
T-RFLP patterns were determined on day 21. In the upper layer, the most dominant T-RFs could be assigned to Dechloromonas (430 to 433 bp), Betaproteobacteria (489 bp), Bacillus (150 to 152 bp), Clostridium (270 to 271 and 515 to 518 bp), and Clostridium cluster I (507 to 510 bp). Betaproteobacteria, including Dechloromonas, had a higher Ap in treatment B microcosms than in treatment P+B and control microcosms, while bacilli and clostridia were more important in the presence of protists. T-RFs assigned to Chloroflexi (120 bp) and Bacillus/Geobacter (128 bp) had also a higher Ap in treatment P+B microcosms but did not occur in all three replicates. Summarizing the patterns for the upper layer, treatment P+B and control microcosms showed greater complexity than treatment B microcosms. For details see the supplemental material.
In the lower layer, the dominant T-RFs were 270- and 510-bp T-RFs, followed by 515-, 145- (assigned to Bacillus), and 152-bp T-RFs. The patterns for the lower layer changed between days 0 and 7, but they stayed quite constant later (see the supplemental material). The most obvious difference between treatments was the difference in the Ap of T-RF 270 (clostridia), which was the dominant T-RF in treatment P+B.
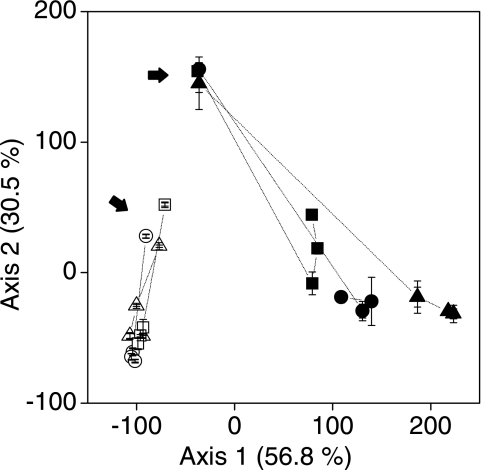
A correspondence analysis of the T-RFLP patterns showed that there was clear separation of samples according to layer, treatment, and time of incubation (Fig. 7). As soon as after 10 h (day 0) the upper and lower layers were clearly separated. In the upper layer, the T-RFLP patterns had changed remarkably after 7 days, and all the treatments were well separated from each other. Treatment P+B was located halfway between treatment B and the control. However, the distance to treatment B increased with time (Fig. 7), while the bacterial diversity increased in the microcosms in which protists were present (treatment P+B and the control) (see the supplemental material). In the lower layer, only minor temporal changes were observed after day 7. We also compared the T-RFLP patterns of the archaeal 16S rRNA gene but found no difference among the treatments (primer Ar 109f and primer Ar 915r labeled with 6-carboxyfluorescein [13] (data not shown).
FIG. 7.
Correspondence analysis of bacterial communities based on T-RFLP patterns. Circles, sterilized soil inoculated with protists and bacteria (treatment P+B); triangles, sterilized soil inoculated with bacteria (treatment B); squares, control. Solid symbols, upper layer; open symbols, lower layer. Day 0 is indicated by arrows, and data from the same treatments are connected with lines in the order of time series. The values are means ± standard errors (n = 3).
DISCUSSION
In spite of the intense biogeochemical, microbiological, and molecular work done on rice fields, the impact of protistan grazing on the structure and function of the prokaryotic community was largely unknown previously. Our results demonstrate effects on different levels, including biomass, population structure, and activities. Without protists, the biomass (total DNA) increased (Fig. 5), Beataproteobacteria instead of bacilli and clostridia dominated the bacterial community (see the supplemental material), the level of extractable NH4+ was lower, and the level of extractable SO42− was higher (Fig. 3 and 4). These effects of protists were most obvious in the partially oxic surface layer (0 to 3 mm) but could also be detected in the totally anoxic subsurface layer (10 to 13 mm).
Experimental design.
Microcosms and microbial model systems in particular allow a degree of control, replication, and reproducibility that is unsurpassed in field experiments (35). Nevertheless, microcosms have been criticized as systems that are too simplistic for certain questions in community or ecosystem ecology (10). However, in a previous study we observed very similar dynamics of and controls on methane-oxidizing bacteria in microcosms and in the field (23). Even if methanotrophs are only a subset of the microbial community, the degree of agreement allowed us to suggest that the microcosms used here are a reliable model for the interaction between protists and bacteria at the soil surface.
Our conclusions depend on effective sterilization of the soil used for the microcosms manipulated. In a recent meta-analysis a γ-radiation dose of 25 kGy was found to be sufficient to kill nearly everything except some radioresistant bacteria (48). The latter are not to be expected to play any role in this rice field soil. Indeed, the sterilized soil produced neither CO2 nor CH4 when it was slurried and incubated anaerobically (data not shown). This indicates that the activities measured for treatments P+B and B were catalyzed by microorganisms from the inocula. Nevertheless, DNA extracted from the sterilized soil could be amplified with eukaryotic primers (Fig. 6, lanes S). The bands could be affiliated with green algae (Fig. 6A and Table 2). However, green algae are sensitive to γ-radiation (48). In addition, the microcosms were incubated in the dark, which prevented growth of phototrophic organisms. Hence, the sequences were thought not to represent live organisms. Because we prepared bacterial inocula by filtering them through a 3-μm filter, some small protists may have passed through. However, only the ubiquitous DGGE bands affiliated with algae were detected in treatment B (Fig. 6A and Table 2), indicating that there was successful exclusion of living protists.
The soil microcosms were not flooded but were water saturated. Using this technique, we could ascertain that the redox gradients typical for flooded soils developed without the growth of free-swimming fauna and flora in the overlying water that might have compromised the analysis of soil-specific interactions (57). Indeed, the oxygen profiles measured in the control microcosms were quite similar to those reported for previous experiments with flooded soil microcosms (44, 51). This suggests that redox conditions typically found in a flooded rice field soil were successfully established in the microcosms.
Biogeochemistry.
After 10 h (day 0), the O2 gradients in the control microcosms were much steeper than those in the treatment B and P+B microcosms (Fig. 1). Similarly, the NO3− content in the control microcosms was below the detection limit (0.01 μmol · g [dry weight] soil−1) on day 0, while the treatment B and P+B microcosms contained 0.7 to 1.1 μmol NO3− · g (dry weight) soil−1. This indicates that the sequential reduction of electron acceptors (45, 54) was retarded in the reinoculated microcosms. Until day 7 the differences became less pronounced, and at day 14 CO2 emission and O2 uptake were highest in treatment P+B microcosms (Fig. 1). This may have been a side effect of γ-radiation, which is reported to facilitate substrate availability in sterilized soils (2). The measured rates were consistent with the depth of penetration of O2 (Fig. 1 and 2). Altogether, this indicates that protists have a stimulating effect on carbon and nitrogen mineralization in water-saturated soils, as found previously for upland soils (1, 14, 22, 27, 53).
The high SO42− concentration in treatment B microcosms (Fig. 4) suggests that protists may also stimulate SO42− reduction. To our knowledge, this is the first evidence for an effect of protists on the sulfur cycle. It is not clear, however, if the protists affect SO42− reduction directly by grazing on SO42−-reducing bacteria or indirectly via creating more reduced soil conditions by stimulating the overall microbial activities.
The level of soil dsDNA correlates well with soil microbial biomass (6, 46). In the upper layer, the total level of dsDNA was lower in the presence of protists than in the absence of protists (Fig. 5), suggesting that protists controlled the microbial population. Different studies have demonstrated that protistan grazing may reduce the number (30, 31, 53) and biomass of bacteria (1). One may argue that part of the dsDNA was extracted from eukaryotes or even from dead biomass. However, the dsDNA content increased with time in all treatments and was highest without protists (Fig. 5). In summary, these findings provide evidence that the difference was due to prokaryotes alone. Based on these assumptions, the microbial biomass in the upper layer in treatment P+B microcosms was one-half that in treatment B microcosms at day 21 (Fig. 5). A similar effect was reported for experiments performed with upland soil microcosms (41). The real impact of protists on microbial biomass may have been even more significant in the oxic surface soil, because (i) the microbial biomass in the lower anoxic layer was apparently not affected by protists (Fig. 5B) and (ii) about one-half of the upper layer was anoxic (Fig. 2) on days 7 to 21.
This estimate was based on the standing stock. However, the smaller biomass in treatment P+B microcosms may have had higher gross production, as indicated by higher activities. Grazing may accelerate nutrient cycling in general (9, 15), and elevated specific metabolic activities are typical for a bacterial population under grazing pressure (1, 3, 41). This effect could also be detected in the anoxic lower layer, even if a eukaryotic community was barely detectable by PCR-DGGE analysis (see below). While the total dsDNA content was not affected, the level of nitrogen mineralization was higher and the level of sulfate reduction was lower in the presence of protists, as indicated by the extractable soil NH4+ and SO42− contents, respectively (Fig. 3 and 4).
Molecular ecology.
T-RFLP and DGGE offer the best compromise between processing time and information gained if a large number of samples has to be processed. For the bacteria, we could rely on a clone library generated previously from the same soil (51). However, in previous work on protists from the soil we focused on ciliates and relied on morphology-based identification (57, 58). To cover the full diversity of protists, we had to use a molecular approach. We opted for an 18S rRNA gene-based assay in combination with DGGE that covered a very wide range of eukaryotic phylotypes (20).
(i) Eukaryotic community.
The majority of the DGGE bands could be affiliated with flagellates. Most of the flagellate-related sequences were assigned to the Cercomonadida in the phylum Cercozoa and to the Cercomonadidae and Heteromitidae in particular (11, 50). Members of the Cercomonadida have been known for a long time as the most abundant and widespread soil flagellates (50), and Cercomonas and Heteromita are common soil bacterivores that graze on attached bacteria (25, 47). They have been shown to affect the bacterial community in vitro (53) and to excrete ammonia as a main form of nitrogen when they are grazing on bacteria (18).
In contrast to the upper layer, the lower layer contained a much less diverse eukaryotic community, with only one sequence assigned to protists (Cercomonas) (Table 2). In a previous experiment, we quantified anaerobic ciliates (direct counts, up to 60 cells · g [dry weight] soil−1) and flagellates (most probable number, up to 700 cells · g [dry weight] soil−1) in the same soil (58), but the resulting number of targets was obviously too low to be detected in the presence of the background algal DNA (Table 2 and Fig. 6C). Less microbial biomass and the low cell yield of anoxic protists (24) may be the factors limiting the development of anaerobic eukaryotes. The only DGGE band affiliated with Cercomonas was obtained from both the control soil and treatment P+B microcosms (Table 2 and Fig. 6C), suggesting that this genus is the dominant genus in anoxic soil also.
(ii) Prokaryotic community.
As soon as 10 h after flooding (day 0) we detected similar bacterial communities in the upper soil layers of the microcosms that received different treatments, indicating that bacterial diversity had been successfully reestablished (Fig. 7). The bacterial communities in upper and lower layers were different (Fig. 7). This finding is in accordance with previous work (51) and correlates well with the rapid evolution of oxygen gradients (Fig. 2). Hence, the availability of e− acceptors is the most probable control during early development of the microbial community. There were no major differences in the microbial succession (days 7 to 21) in the lower layer between treatments (Fig. 7). In the upper layer, the bacterial community changed much more with time. The bacterial communities in the treatment P+B and control microcosms were not identical, but they were more similar to each other than to the bacterial communities in the treatment B microcosms, at least at day 21 (Fig. 7). The archaeal community, however, stayed constant. The most conspicuous change in the bacterial community was the high abundance of T-RFs affiliated with Betaproteobacteria (430 and 489 bp) in treatment B. In the presence of protists, low-G+C-content gram-positive bacteria (Bacillus and clostridia) were more dominant. This may suggest that Betaproteobacteria were grazed upon preferably (see the supplemental material).
The upper layer as defined and sampled in this experiment covers the oxic-anoxic boundary zone. Many important redox reactions take place at this highly active interface, including methane oxidation, oxidation of reduced metal ions, coupled nitrification-denitrification, sulfurication, and other reactions. In this paper we show the gross effects of grazing. It will be challenging to become more specific and to study the effect of protistan grazing on particular processes in detail, but this is a promising area of research.
Supplementary Material
Acknowledgments
We thank R. Conrad and M. V. J. Schwarz for helpful hints and comments and S. Russo, Istituto Sperimentale per la Ceriocultura, Vercelli, Italy, for providing soil samples.
This study was supported by a grant from the Deutsche Forschungs Gemeinschaft (SFB 395, “Interaction, Adaptation and Catalytic Capabilities of Microorganisms in Soil”). J. Murase was supported by the Ministry of Education, Sports, Science, Culture, and Technology, Japan.
Footnotes
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Alphei, J., M. Bonkowski, and S. Scheu. 1996. Protozoa, Nematoda and Lumbricidae in the rhizosphere of Hordelymus europaeus (Poaceae)—faunal interactions, response of microorganisms and effects on plant growth. Oecologia 106:111-126. [DOI] [PubMed] [Google Scholar]
- 2.Alphei, J., and S. Scheu. 1993. Effects of biocidal treatments on biological and nutritional properties of a mull-structured woodland soil. Geoderma 56:435-445. [Google Scholar]
- 3.Anderson, R. V., E. T. Elliott, J. F. Mcclellan, D. C. Coleman, C. V. Cole, and H. W. Hunt. 1978. Trophic interactions in soils as they affect energy and nutrient dynamics. 3. Biotic interactions of bacteria, amebas, and nematodes. Microb. Ecol. 4:361-371. [DOI] [PubMed] [Google Scholar]
- 4.Badalucco, L., F. Pomare, S. Grego, L. Landi, and P. Nannipieri. 1994. Activity and degradation of streptomycin and cycloheximide in soil. Biol. Fertil. Soils 18:334-340. [Google Scholar]
- 5.Bak, F., G. Scheff, and K. H. Jansen. 1991. A rapid and sensitive ion chromatographic technique for the determination of sulfate and sulfate reduction rates in freshwater lake sediments. FEMS Microbiol. Ecol. 85:23-30. [Google Scholar]
- 6.Blagodatskaya, E. V., S. A. Blagodatskii, and T. H. Anderson. 2003. Quantitative isolation of microbial DNA from different types of soils of natural and agricultural ecosystems. Microbiology 72:744-749. [PubMed] [Google Scholar]
- 7.Bodelier, P. L. E. 2003. Interactions between oxygen-releasing roots and microbial processes in flooded soils and sediments, p. 331-362. In H. de Kroon and E. J. W. Visser (ed.), Root ecology. Springer, Berlin, Germany.
- 8.Bodelier, P. L. E., P. Roslev, T. Henckel, and P. Frenzel. 2000. Stimulation by ammonium-based fertilizers of methane oxidation in soil around rice roots. Nature 403:421-424. [DOI] [PubMed] [Google Scholar]
- 9.Bonkowski, M. 2004. Protozoa and plant growth: the microbial loop in soil revisited. New Phytol. 162:617-631. [DOI] [PubMed] [Google Scholar]
- 10.Carpenter, S. R. 1996. Microcosm experiments have limited relevance for community and ecosystem ecology. Ecology 77:677-680. [Google Scholar]
- 11.Cavalier-Smith, T., and E. E. Y. Chao. 2003. Phylogeny and classification of phylum Cercozoa (Protozoa). Protist 154:341-358. [DOI] [PubMed] [Google Scholar]
- 12.Chakraborty, N., G. M. Sarkar, and S. C. Lahiri. 2003. Effect of physical irradiation and chemical mutagen treatment on methane production by methanogenic bacteria. World J. Microbiol. Biotechnol. 19:145-150. [Google Scholar]
- 13.Chin, K. J., T. Lukow, and R. Conrad. 1999. Effect of temperature on structure and function of the methanogenic archaeal community in an anoxic rice field soil. Appl. Environ. Microbiol. 65:2341-2349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Clarholm, M. 1985. Interactions of bacteria, protozoa and plants leading to mineralization of soil-nitrogen. Soil Biol. Biochem. 17:181-187. [Google Scholar]
- 15.Clarholm, M. 2002. Bacteria and protozoa as integral components of the forest ecosystem and their role in creating a naturally varied soil fertility. Antonie Leeuwenhoek 81:309-318. [DOI] [PubMed] [Google Scholar]
- 16.Conrad, R., and P. Frenzel. 2002. Flooded soils, p. 1316-1333. In G. Britton (ed.), Encyclopedia of environmental microbiology. John Wiley & Sons, New York, N.Y.
- 17.Corbin, J. L. 1984. Liquid-chromatographic fluorescence determination of ammonia from nitrogenase reactions: a 2-min assay. Appl. Environ. Microbiol. 47:1027-1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Darbyshire, J. F., K. B. Zwart, and D. A. Elston. 1993. Growth and nitrogenous excretion of a common soil flagellate, Cercomonas sp. Soil Biol. Biochem. 25:1583-1589. [Google Scholar]
- 19.del Giorgio, P. A., J. M. Gasol, D. Vaqué, P. Mura, S. Agustí, and C. M. Duarte. 1996. Bacterioplankton community structure: protists control net production and the proportion of active bacteria in a coastal marine community. Limnol. Oceanogr. 41:1169-1179. [Google Scholar]
- 20.Díez, B., C. Pedrós-Alió, T. L. Marsh, and R. Massana. 2001. Application of denaturing gradient gel electrophoresis (DGGE) to study the diversity of marine picoeukaryotic assemblages and comparison of DGGE with other molecular techniques. Appl. Environ. Microbiol. 67:2942-2951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Edwards, U., T. Rogall, H. Blöcker, M. Emde, and E. C. Böttger. 1989. Isolation and direct complete determination of entire genes. Nucleic Acids Res. 17:7843-7853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ekelund, F., and R. Rønn. 1994. Notes on protozoa in agricultural soil with emphasis on heterotrophic flagellates and naked amoebae and their ecology. FEMS Microbiol. Rev. 15:321-353. [DOI] [PubMed] [Google Scholar]
- 23.Eller, G., M. Krüger, and P. Frenzel. 2005. Comparing field and microcosm experiments: a case study on methano- and methylotrophic bacteria in paddy soil. FEMS Microbiol. Ecol. 51:279-291. [DOI] [PubMed] [Google Scholar]
- 24.Fenchel, T., and B. J. Finlay. 1995. Ecology and evolution in anoxic worlds. Oxford University Press, Oxford, United Kingdom.
- 25.Fredslund, L., F. Ekelund, C. S. Jacobsen, and K. Johnsen. 2001. Development and application of a most-probable-number-PCR assay to quantify flagellate populations in soil samples. Appl. Environ. Microbiol. 67:1613-1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Goyal, S. S., D. W. Rains, and R. C. Huffaker. 1988. Determination of ammonium ion by fluorometry or spectrophotometry after online derivatization with ortho-phthalaldehyde. Anal. Chem. 60:175-179. [DOI] [PubMed] [Google Scholar]
- 27.Griffiths, B. S., and I. M. Young. 1994. The effects of soil structure on protozoa in a clay-loam soil. Eur. J. Soil Sci. 45:285-292. [Google Scholar]
- 28.Grosse, W., J. Armstrong, and W. Armstrong. 1996. A history of pressurised gas-flow studies in plants. Aquat. Bot. 54:87-100. [Google Scholar]
- 29.Grosskopf, R., P. H. Janssen, and W. Liesack. 1998. Diversity and structure of the methanogenic community in anoxic rice paddy soil microcosms as examined by cultivation and direct 16S rRNA gene sequence retrieval. Appl. Environ. Microbiol. 64:960-969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Habte, M., and M. Alexander. 1975. Protozoa as agents responsible for decline of Xanthomonas campestris in soil. Appl. Microbiol. 29:159-164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Habte, M., and M. Alexander. 1977. Further evidence for the regulation of bacterial populations in soil by protozoa. Arch. Microbiol. 113:181-183. [DOI] [PubMed] [Google Scholar]
- 32.Hahn, M. W., and M. G. Höfle. 2001. Grazing of protozoa and its effect on populations of aquatic bacteria. FEMS Microbiol. Ecol. 35:113-121. [DOI] [PubMed] [Google Scholar]
- 33.Holzapfel-Pschorn, A., R. Conrad, and W. Seiler. 1986. Effects of vegetation on the emission of methane from submerged paddy soil. Plant Soil 92:223-233. [Google Scholar]
- 34.Jackson, M. B., and W. Armstrong. 1999. Formation of aerenchyma and the processes of plant ventilation in relation to soil flooding and submergence. Plant Biol. 1:274-287. [Google Scholar]
- 35.Jessup, C. M., R. Kassen, S. E. Forde, B. Kerr, A. Buckling, P. B. Rainey, and B. J. M. Bohannan. 2004. Big questions, small worlds: microbial model systems in ecology. Trends Ecol. Evol. 19:189-197. [DOI] [PubMed] [Google Scholar]
- 36.Jürgens, K., and C. Matz. 2002. Predation as a shaping force for the phenotypic and genotypic composition of planktonic bacteria. Antonie Leeuwenhoek 81:413-434. [DOI] [PubMed] [Google Scholar]
- 37.Kimura, M. 2000. Anaerobic microbiology in waterlogged rice fields. Soil Biochem. 10:35-138. [Google Scholar]
- 38.Kirk, G. 2004. The biogeochemistry of submerged soils. John Wiley & Sons, Ltd., Chichester, United Kingdom.
- 39.Krüger, M., P. Frenzel, and R. Conrad. 2001. Microbial processes influencing methane emission from rice fields. Global Change Biol. 7:49-63. [Google Scholar]
- 40.Krüger, M., P. Frenzel, D. Kemnitz, and R. Conrad. 2005. Activity, structure and dynamics of the methanogenic archaeal community in a flooded Italian rice field. FEMS Microbiol. Ecol. 51:323-331. [DOI] [PubMed] [Google Scholar]
- 41.Kuikman, P. J., A. G. Jansen, J. A. VanVeen, and A. J. B. Zehnder. 1990. Protozoan predation and the turnover of soil organic-carbon and nitrogen in the presence of plants. Biol. Fertil. Soils 10:22-28. [Google Scholar]
- 42.Lane, D. J. 1991. 16S/23S rRNA sequencing, p. 115-175. In E. Stackebrandt and M. Goodfellow (ed.), Nucleic acid techniques in bacterial systematics. John Wiley & Sons Ltd., Chichester, England.
- 43.Liesack, W., S. Schnell, and N. P. Revsbech. 2000. Microbiology of flooded rice paddies. FEMS Microbiol. Rev. 24:625-645. [DOI] [PubMed] [Google Scholar]
- 44.Lüdemann, H., I. Arth, and W. Liesack. 2000. Spatial changes in the bacterial community structure along a vertical oxygen gradient in flooded paddy soil cores. Appl. Environ. Microbiol. 66:754-762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lueders, T., and M. Friedrich. 2000. Archaeal population dynamics during sequential reduction processes in rice field soil. Appl. Environ. Microbiol. 66:2732-2742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Marstorp, H., and E. Witter. 1999. Extractable dsDNA and product formation as measures of microbial growth in soil upon substrate addition. Soil Biol. Biochem. 31:1443-1453. [Google Scholar]
- 47.Mattison, R. G., H. Taki, and S. Harayama. 2002. The bacterivorous soil flagellate Heteromita globosa reduces bacterial clogging under denitrifying conditions in sand-filled aquifer columns. Appl. Environ. Microbiol. 68:4539-4545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.McNamara, N. P., H. I. J. Black, N. A. Beresford, and N. R. Parekh. 2003. Effects of acute gamma irradiation on chemical, physical and biological properties of soils. Appl. Soil Ecol. 24:117-132. [Google Scholar]
- 49.Mitsch, W. J., and J. G. Gosselink. 1993. Wetlands. Van Nostrand Reinhold, New York, N.Y.
- 50.Myl'nikov, A. P., and S. A. Karpov. 2004. Review of diversity and taxonomy of cercomonads. Protistology 3:201-217. [Google Scholar]
- 51.Noll, M., D. Matthies, P. Frenzel, M. Derakshani, and W. Liesack. 2005. Succession of bacterial community structure and diversity in a paddy soil oxygen gradient. Environ. Microbiol. 7:382-395. [DOI] [PubMed] [Google Scholar]
- 52.Pernthaler, J., T. Posch, K. Simek, J. Vrba, A. Pernthaler, F. O. Glockner, U. Nubel, R. Psenner, and R. Amann. 2001. Predator-specific enrichment of actinobacteria from a cosmopolitan freshwater clade in mixed continuous culture. Appl. Environ. Microbiol. 67:2145-2155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rønn, R., A. E. McCaig, B. S. Griffiths, and J. I. Prosser. 2002. Impact of protozoan grazing on bacterial community structure in soil microcosms. Appl. Environ. Microbiol. 68:6094-6105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Roy, R., H. D. Klüber, and R. Conrad. 1997. Early initiation of methane production in anoxic rice soil despite the presence of oxidants. FEMS Microbiol. Ecol. 24:311-320. [Google Scholar]
- 55.Sanders, R. W., and K. G. Porter. 1986. Use of metabolic inhibitors to estimate protozooplankton grazing and bacterial production in a monomictic eutrophic lake with an anaerobic hypolimnion. Appl. Environ. Microbiol. 52:101-107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Scheid, D., S. Stubner, and R. Conrad. 2004. Identification of rice root associated nitrate, sulfate and ferric iron reducing bacteria during root decomposition. FEMS Microbiol. Ecol. 50:101-110. [DOI] [PubMed] [Google Scholar]
- 57.Schwarz, M. V. J., and P. Frenzel. 2003. Population dynamics and ecology of ciliates (Protozoa, Ciliophora) in an anoxic rice field soil. Biol. Fertil. Soils 38:245-252. [Google Scholar]
- 58.Schwarz, M. V. J., and P. Frenzel. 2005. Methanogenic symbionts of anaerobic ciliates and their contribution to methanogenesis in an anoxic rice field soil. FEMS Microbiol. Ecol. 52:93-99. [DOI] [PubMed] [Google Scholar]
- 59.Tremaine, S. C., and A. L. Mills. 1987. Inadequacy of the eukaryote inhibitor cycloheximide in studies of protozoan grazing on bacteria at the freshwater-sediment interface. Appl. Environ. Microbiol. 53:1969-1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Weber, S., S. Stubner, and R. Conrad. 2001. Bacterial populations colonizing and degrading rice straw in anoxic paddy soil. Appl. Environ. Microbiol. 67:1318-1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.