Abstract
A gram-positive strong polychlorinated biphenyl (PCB) degrader, Rhodococcus sp. strain RHA1, can degrade PCBs by cometabolism with biphenyl or ethylbenzene. In RHA1, three sets of aromatic-ring-hydroxylating dioxygenase genes are induced by biphenyl. The large and small subunits of their terminal dioxygenase components are encoded by bphA1 and bphA2, etbA1 and etbA2, and ebdA1 and ebdA2, respectively, and the deduced amino acid sequences of etbA1 and etbA2 are identical to those of ebdA1 and ebdA2, respectively. In this study, we examined the involvement of the respective subunit genes in biphenyl/PCB degradation by RHA1. Reverse transcription-PCR and two-dimensional polyacrylamide gel electrophoresis analyses indicated the induction of RNA and protein products of etbA1 and ebdA1 by biphenyl. Single- and double-disruption mutants of etbA1, ebdA1, and bphA1 were constructed by insertional inactivation. The 4-chlorobiphenyl (4-CB) degradation activities of all the mutants were lower than that of RHA1. The results indicated that all of these genes are involved in biphenyl/PCB degradation. Furthermore, we constructed disruption mutants of ebdA3 and bphA3, encoding ferredoxin, and etbA4, encoding ferredoxin reductase components. The 4-CB degradation activities of these mutants were also lower than that of RHA1, suggesting that all of these genes play a role in biphenyl/PCB degradation. The substrate preferences of etbA1A2/ebdA1A2- and bphA1A2-encoded dioxygenases for PCB congeners were examined using the corresponding mutants. The results indicated that these dioxygenase isozymes have different substrate preferences and that the etbA1A2/ebdA1A2-encoded isozyme is more active on highly chlorinated congeners than the bphA1A2-encoded one.
Polychlorinated biphenyls (PCBs) are synthetic compounds that have excellent chemical stability, insulation properties, and resistance to combustion, and they have been used in a wide range of industrial applications. However, since the revelation that PCBs have strong toxicity and are recalcitrant in the environment, environmental contamination by PCBs has become a serious worldwide problem. Degradation of PCBs by microorganisms has been regarded as an effective tool for removal of PCBs from the environment, and many kinds of bacteria which degrade PCBs aerobically have been isolated and characterized (2, 4, 7, 11, 14). These bacteria degrade PCBs by cometabolism, with PCBs being degraded via the biphenyl catabolic pathway in the presence of biphenyl (5, 10). In the initial step of the biphenyl catabolic pathway, biphenyl is transformed to 2,3-dihydroxy-1-phenylcyclohexa-4,6-diene (dihydrodiol compound) by an aromatic-ring-hydroxylating dioxygenase (RHDO), biphenyl 2,3-dioxygenase (BDO) (Fig. 1A), which consists of the large and small subunits of terminal oxygenase, ferredoxin, and ferredoxin reductase. BDO is a key enzyme that primarily determines the substrate specificity of PCB degradation.
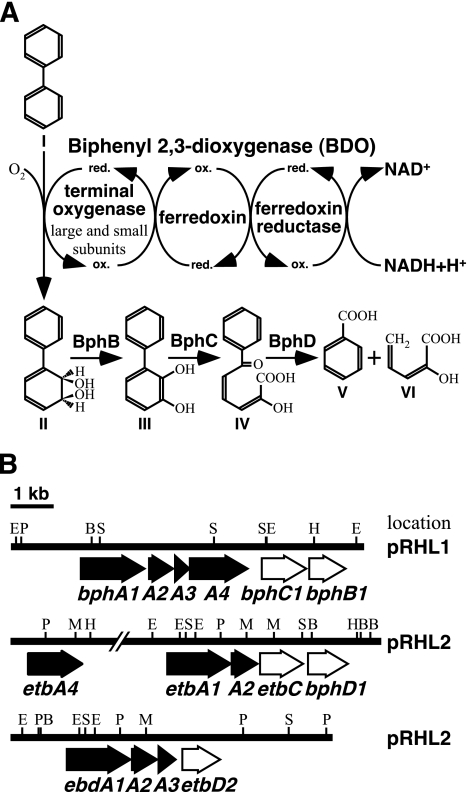
FIG. 1.
Components and functions of BDO and proposed pathway for aerobic degradation of biphenyl (A) and organization of the bph, etb, and ebd genes (B) in Rhodococcus sp. strain RHA1. (A) The proposed electron transfer reactions and the conversion of biphenyl to benzoate and 2-hydroxypenta-2,4-dienoate are indicated. The BDO consists of three components, including a terminal oxygenase complex of large and small subunits, a ferredoxin, and a ferredoxin reductase. The complex catalyzes the dihydroxylation of biphenyl and is preferentially involved in the determination of substrate specificity. Ferredoxin and ferredoxin reductase promote electron transfer from NADH to the large- and small-subunit complex. Compounds: I, biphenyl; II, cis-(2R,3S)-dihydroxy-1-phenylcyclohexa-4,6-diene (dihydrodiol); III, 2,3-dihydroxybiphenyl (2,3-DHBP); IV, 2-hydroxy-6-oxo-6-phenylhexa-2,4-dienoate (meta-cleavage compound [HOPD]); V, benzoate; VI, 2-hydroxypenta-2,4-dienoate. Enzymes: BphB, dihydrodiol dehydrogenase; BphC, 2,3-DHBP 1,2-dioxygenase; BphD, HOPD hydrolase. (B) Arrows indicate ORFs. The filled arrows represent the ORFs encoding BDO subunits. The names of the genes are indicated below the arrows. The etbA4 gene is located 6.0 kb upstream from etbA1. The bphA1, ebdA1, and etbA1 genes encode the large subunit of BDO, the bphA2, ebdA2, and etbA2 genes encode the small subunit of BDO, bphA3 and ebdA3 encode the ferredoxin, and bphA4 and etbA4 encode the ferredoxin reductase. The plasmid location of each gene segment is indicated on the right. Abbreviations: B, BamHI; E, EcoRI; H, HindIII; M, MluI; P, PstI; S, StuI.
Rhodococcus sp. strain RHA1 is a gram-positive strong PCB degrader. RHA1 can degrade mono- to octachlorobiphenyls by cometabolism with biphenyl (17). Various genes responsible for biphenyl and PCB degradation in RHA1 have been isolated and characterized (8, 12, 13, 15, 19, 20). The bphA1A2A3A4 genes encoding BDO were initially isolated from and characterized for RHA1 (13). Based on results from PCR amplification of RHDO genes using consensus primers and Northern hybridization analysis with the amplified fragments as probes, Kitagawa et al. indicated that three RHDO large-subunit genes are induced by biphenyl in RHA1 (9). These three genes are bphA1 and two other genes. The latter two genes share an identical nucleotide sequence except for a single nucleotide, and their deduced amino acid sequences are identical. These genes were expected to be involved in the PCB degradation induced by ethylbenzene in RHA1 and RDA1, which is a bphA1 insertion mutant of RHA1 (16), and they were designated etbA1 and ebdA1 (18). The nucleotide sequences of the flanking regions of these genes were determined, and it was revealed that the genes are included in the gene clusters shown in Fig. 1B. Open reading frames (ORFs) downstream of etbA1 and ebdA1, which encode RHDO small subunits, were designated etbA2 and ebdA2, respectively. Their nucleotide sequences are completely identical. Furthermore, an ORF which showed similarity to that for ferredoxin was found downstream from ebdA1A2 and was designated ebdA3. The ebdA1A2A3 cluster is located 6 kb downstream from etbA1A2. In the region located 6 kb upstream from etbA1A2, an ORF whose gene product exhibited 34.2% identity with that of bphA4 was found and was designated etbA4. RHA1 has three linear plasmids, pRHL1 (1,100 kb), pRHL2 (450 kb), and pRHL3 (330 kb). The bphA1A2A3A4 gene cluster is located on pRHL1, and the etbA4, etbA1A2, and ebdA1A2A3 genes are located on pRHL2 (12, 18). Recently, Takeda et al. indicated that the gene clusters including bphA1, etbA1, ebdA1, and etbA4 are under the control of the bphST-encoded two-component regulatory system and are inducibly transcribed in the presence of biphenyl (18). The BphST system is also responsible for transcriptional induction by ethylbenzene, toluene, and other aromatic compounds (19).
In this paper, we examine the involvement of the RHDO genes in the biphenyl/PCB degradation activity of RHA1. We constructed single- and double-disruption mutants of RHDO large-subunit genes and investigated the 4-chlorobiphenyl (4-CB) degradation activities of these mutants. In addition, we constructed insertion mutants of electron transfer component genes. Our results revealed that all of these genes are involved in biphenyl/PCB degradation activity in RHA1.
MATERIALS AND METHODS
Bacterial strains, plasmids, and culture conditions.
The bacterial strains and plasmids used in this study are listed in Table 1. Vector plasmids pFAJ2574 (3), pK4 (6), and pBsRG6 (8) were used as sources for plasmid construction. Rhodococcus strains were grown in Luria-Bertani (LB) medium, 1/5 LB medium (2 g of Bacto tryptone, 1 g of Bacto yeast extract, 5 g of NaCl per liter), 1/3 LB medium (3.33 g of Bacto tryptone, 1.66 g of Bacto yeast extract, 5 g of NaCl per liter), or W minimal medium (13) with biphenyl or vapor of ethylbenzene at 30°C. Escherichia coli JM109 was used as a host strain and was grown in LB medium at 37°C.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Relevant characteristic(s)a | Reference |
|---|---|---|
| Strains | ||
| Rhodococcus sp. strain RHA1 | Wild type; BPH+ ETB+ | 17 |
| Rhodococcus sp. strain HDT1 | Mutant derivative of RHA1; aphII gene insertion mutant of etbA1; Kmr | This study |
| Rhodococcus sp. strain HDB1 | Mutant derivative of RHA1; aphII gene insertion mutant of ebdA1; Kmr | This study |
| Rhodococcus sp. strain HDA1 | Mutant derivative of RHA1; aphII gene insertion mutant of bphA1; Kmr | This study |
| Rhodococcus sp. strain HDB3 | Mutant derivative of RHA1; aphII gene insertion mutant of ebdA3; Kmr | This study |
| Rhodococcus sp. strain HDA3 | Mutant derivative of RHA1; aphII gene insertion mutant of bphA3; Kmr | This study |
| Rhodococcus sp. strain HDT4 | Mutant derivative of RHA1; aphII gene insertion mutant of etbA4; Kmr | This study |
| Rhodococcus sp. strain HDAT1 | Mutant derivative of HDA1; cmrA gene insertion mutant of etbA1; Kmr Cmr | This study |
| Rhodococcus sp. strain HDAB1 | Mutant derivative of HDA1; cmrA gene insertion mutant of ebdA1; Kmr Cmr | This study |
| Rhodococcus sp. strain HDBT1 | Mutant derivative of HDB1; cmrA gene insertion mutant of etbA1; Kmr Cmr | This study |
| Rhodococcus sp. strain HDAB33 | Mutant derivative of HDA3; cmrA gene insertion mutant of ebdA3; Kmr Cmr | This study |
| Escherichia coli JM109 | recA1 endA1 gyrA96 thi-1 hsdR17(rK− mK−) e14 (mcrA) supE44 relA1 Δ(lac-proAB) [F′ traD36 proAB lacIqZΔM15] | |
| Plasmids | ||
| pBluescript II SK(+) | Cloning vector; Apr | Stratagene |
| pETQ1 | pUC119 with 10.0-kb HindIII fragment of RHA1 carrying etbA1A2C and bphD1 | This study |
| pHX11 | pUC119 with 3.8-kb XhoI fragment of RHA1 carrying ebdA1A2A3 | This study |
| pK4tsr | pK4 with insertion of tsr gene from pBsRG6; Kmr Tsr | This study |
| pBlueKm | pBluescript II SK(+) with insertion of aphII gene; Apr Kmr | This study |
| pBKE2 | pBlueKm containing a 761-bp StuI-PvuII etbA1 internal fragment | This study |
| pBAK1 | pBlueKm containing a 1,099-bp Eco52I bphA1 internal fragment | This study |
| pBKEA3 | pBlueKm containing a 330-bp AatII-NarI ebdA3 internal fragment | This study |
| pBKBA3 | pBlueKm containing a 302-bp PCR-amplified bphA3 internal fragment | This study |
| pBKA4-2 | pBlueKm containing a 731-bp MluI-SmaI etbA4 internal fragment | This study |
| pBlueCm | pBluescript II SK(+) with insertion of cmrA gene; Apr Cmr | This study |
| pBCEA1 | pBlueCm containing a 761-bp StuI-PvuII etbA1 internal fragment | This study |
| pBCEB3 | pBlueCm containing a 330-bp AatII-NarI ebdA3 internal fragment | This study |
| pK4TEB1 | pK4tsr with 2.3-kb BamHI-MluI fragment of RHA1 carrying ebdA1 | This study |
| pK4TEB3 | pK4tsr with 1.0-kb EcoRV-KpnI fragment of RHA1 carrying ebdA3 | This study |
| pK4TBA21 | pK4tsr with 2.7-kb EcoRI-NspV fragment of RHA1 carrying bphA1 | This study |
| pK4TBA3 | pK4tsr with 0.5-kb NspV-StuI fragment of RHA1 carrying bphA3 | This study |
| pK4TBA4-2 | pK4tsr with 2.0-kb BglII-HindIII fragment of RHA1 carrying etbA4 | This study |
BPH+, growth on biphenyl; ETB+, growth on ethylbenzene; Apr, ampicillin resistance; Cmr, chloramphenicol resistance; Kmr, kanamycin resistance; Tsr, thiostrepton resistance.
RT-PCR.
Total RNA of RHA1 was prepared from cells grown on biphenyl, ethylbenzene, or LB medium as described by Ausubel et al. (1). Reverse transcription-PCR (RT-PCR) was carried out using a BcaBest RNA PCR kit, version 1.1 (Takara Shuzo, Kyoto, Japan), according to the manufacturer's instructions. Forward primers were designed using the sequences 32 to 55 bp and 29 to 46 bp upstream from the etbA1 and ebdA1 start codons, respectively. These sequences are located downstream from the transcriptional start site of each gene (18). The etbA1 forward, ebdA1 forward, and common reverse primer sequences were 5′-CGTTCACAAATAGTCGCAATTACG-3′, 5′-GACCATTTGCCGTGCTGG-3′, and 5′-CGATGCGGACAAGAATTCG-3′, respectively. The expected sizes of the PCR products for etbA1 and ebdA1 were 330 bp and 321 bp, respectively. To check that the primer pair for etbA1 or ebdA1 did not amplify the other gene, pETQ1 and pHX11, carrying etbA1 and ebdA1, respectively, were used as template DNAs.
2-D PAGE.
RHA1 was grown in 500 ml of 1/5 LB medium to an optical density at 600 nm (OD600) of approximately 1.5. The cells were harvested by centrifugation, washed twice with 5 ml of W minimal medium, and resuspended in W minimal medium to give an OD600 of 1.5. The cell suspension was incubated for 16 h with biphenyl at 30°C. The cells were then harvested by centrifugation, washed twice with 5 ml of 25 mM potassium phosphate buffer (pH 7.0), and resuspended in 10 ml of 25 mM potassium phosphate buffer (pH 7.0). They were disrupted by a single passage through a French press (SLM-Aminco, Rochester, NY) at an operating pressure of 20,000 lb/in2. Cell debris was removed by centrifugation (30,000 × g, 4°C, 15 min). The clear supernatant fluid was used for two-dimensional polyacrylamide gel electrophoresis (2D-PAGE) as a protein sample. The concentration of protein in a sample was determined by means of a protein assay kit (Bio-Rad Laboratories, Hercules, Calif.), using bovine serum albumin as a standard. 2-D PAGE was carried out using a Mini-PROTEAN II 2-D cell (Bio-Rad Laboratories) according to the instructions of the manufacturer. Proteins (0.06 to 0.255 mg) were separated in the first dimension by isoelectric focusing on a pI gradient ranging from 4 to 7 and then further separated in the second dimension by sodium dodecyl sulfate-PAGE with a 12% polyacrylamide gel. After the second-dimension gel electrophoresis, the proteins were stained with a Coomassie brilliant blue R250 or silver stain kit (ATTO, Tokyo, Japan).
N-terminal amino acid sequencing.
The gel pieces containing target proteins were excised from the Coomassie brilliant blue-stained second-dimension gel and subjected to sodium dodecyl sulfate-PAGE analysis. After electrophoresis, the target proteins were blotted electrically onto a polyvinylidene difluoride membrane at 20 V for 1 h using a Trans-Blot SD cell (Bio-Rad). The part of the polyvinylidene difluoride membrane containing the target protein was excised and applied directly to a PPSQ-23 protein sequencer (Shimadzu, Kyoto, Japan).
Gene disruption.
The strategy for gene disruption in RHA1 was described previously (8). To construct single-insertion mutants, the 761-bp StuI-PvuII, 1,099-bp Eco52I, 330-bp AatII-NarI, and 731-bp MluI-SmaI fragments containing the internal segments of etbA1, bphA1, ebdA3, and etbA4, respectively, were inserted into pBlueKm, which is a pBluescript II SK(+) derivative harboring the aphII gene. The internal segment of bphA3 was amplified using the bphA3-F (5′-CCTCACAAAGATATGCAGCTCCGG-3′) and bphA3-Rs (5′-CCTCTGCTGCAAGAGCACCTGGAGATC-3′) primer pair and was inserted into pBlueKm. The resulting plasmids, pBKE2, pBAK1, pBKEA3, pBKA4-2, and pBKBA3, respectively, were introduced independently into RHA1 cells by electroporation. Transformants were selected on 1/3 LB agar plates containing 50 mg of kanamycin per liter and were subjected to Southern hybridization analysis in order to examine the insertion of pBKE2, pBAK1, pBKEA3, pBKA4-2, and pBKBA3 into etbA1, bphA1, ebdA3, etbA4, and bphA3, respectively, by single crossovers. Because ebdA1 is identical to etbA1, an ebdA1 insertion was selected along with an etbA1 insertion. In an insertion mutant, a pBlueKm segment is expected to be sandwiched between a pair of genes inactivated by terminal deletions.
To construct double-insertion mutants, the 761-bp StuI-PvuII and 330-bp AatII-NarI fragments containing the internal segments of etbA1 and ebdA3, respectively, were inserted into pBlueCm, which is a pBluescript II SK(+) derivative harboring the cmrA gene (3). The resulting plasmids, pBCEA1 and pBCEB3, respectively, were introduced independently into the cells of a single-insertion mutant by electroporation. Transformants were selected on 1/3 LB agar plates containing 50 mg of kanamycin and 15 mg of chloramphenicol per liter and were subjected to Southern hybridization analysis to examine the insertion of pBCEA1 and pBCEB3 into etbA1 or ebdA1 and ebdA3 by single crossover.
Resting cell assay.
RHA1 and its insertion mutants were grown on 1/5 LB medium at 30°C to give an OD600 of 0.2, and then the degradation genes were induced by incubation with vapor of ethylbenzene for another 16 h. Cells were collected by centrifugation, washed twice with 2 ml of W minimal medium, and resuspended in W minimal medium to give an OD600 of 1.0. One milliliter of cell suspension was preincubated for 5 min at 30°C and was incubated in a sealed 4.5-ml glass vial with shaking at 30°C after the addition of 50 nmol of 4-CB or 1 ppm each of PCB congeners. The cell suspensions of single- and double-disruption mutants were incubated for 60 and 120 min, respectively, with 4-CB and for 24 h with PCB congeners. Control cells were inactivated by being autoclaved at 121°C for 15 min prior to the addition of substrate. To stop the reaction, 0.1 ml of 6 N HCl was added to the mix, and 50 nmol of phenanthrene or 5 nmol of 4-chlorobiphenyl was added as an internal standard. After the addition of NaCl to saturation, 3 ml of ethyl acetate was added to the mixture, which was then mixed on a vortex mixer for 1 min. The supernatant was recovered, dehydrated with sodium sulfate, evaporated, and dissolved in 0.1 ml of ethyl acetate. One microliter of extract was analyzed by gas chromatography-mass spectrometry (GC-MS) (model 5971A; Agilent Technologies Co., Palo Alto, Calif.), using an Ultra-2 capillary column (50 m by 0.2 mm; Agilent Technologies Co.) as described previously (17).
RESULTS AND DISCUSSION
Expression of etbA1 and ebdA1 in RHA1.
To discriminate between the etbA1 and ebdA1 transcripts, RT-PCR analysis was performed with RNAs from RHA1 cells grown on biphenyl, ethylbenzene, or LB, using the sequences upstream from etbA1 and ebdA1 as one of the primers, since the sequences upstream from these two genes are different. The etbA1 and ebdA1 forward primers were designed using the sequences downstream from the transcriptional start site of each gene, as determined by Takeda et al. (18). The primer pair for etbA1 did not produce any PCR product with ebdA1 plasmid DNA as the template, and that for ebdA1 produced no PCR product with etbA1 plasmid DNA as the template (data not shown). When total RNAs of the cells grown on biphenyl or ethylbenzene were used as templates, PCR products with the expected sizes for etbA1 and ebdA1 were obtained. No PCR product was obtained from the cells grown in LB medium (data not shown). Takeda et al. indicated the induction of bphA1 gene transcription in the presence of biphenyl or ethylbenzene by RT-PCR (19) and suggested that transcription from five adjacent promoters upstream from the degradation gene clusters containing etbA1, ebdA1, and bphA1 is coordinately controlled by the BphST system (18). The BphST system induced transcription beginning with each of the five promoters in response to ethylbenzene and other aromatic compounds as well as biphenyl (19). Our results indicated that etbA1 and ebdA1 as well as bphA1 are inducibly transcribed in the presence of biphenyl or ethylbenzene.
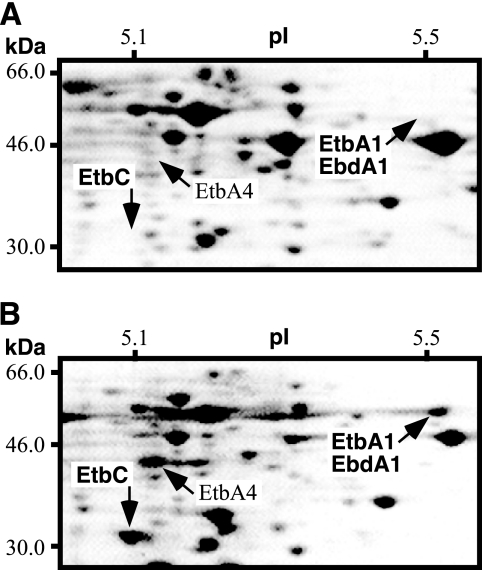
To verify the protein products of etbA1 and ebdA1 in RHA1 induced during incubation with biphenyl, we performed 2-D PAGE (Fig. 2). The results for protein samples from the cells grown on biphenyl revealed the identity of a protein spot whose molecular mass and isoelectric point (pI) were 51 kDa and 5.5, respectively (Fig. 2B). These values were very close to those of 51.7 kDa and 5.2, respectively, which were estimated from the amino acid sequence of EtbA1/EbdA1. The N-terminal amino acid sequence of the protein from this spot was determined, and the obtained sequence, MLXSEXFSPG (X is an unknown), was correspondent with the sequence of EtbA1 and EbdA1 (MLRSERFSPG). These results indicated that either etbA1, ebdA1, or both are inducibly expressed as a protein product in RHA1 grown on biphenyl. In addition to EtbA1/EbdA1, 15 protein spots were unique to the cells grown on biphenyl in comparison to those grown in LB medium. One spot with a molecular mass of 34 kDa and a pI of 4.8 was assigned as the EtbC dioxygenase protein based on its deduced molecular mass, pI, and N-terminal amino acid sequence, AKVTELGYL. Another spot with a molecular mass of 45 kDa and a pI of 5.1 was estimated to be EtbA4, whose deduced molecular mass and pI are 44 kDa and 4.9, respectively (Fig. 2B).
FIG. 2.
Two-dimensional polyacrylamide gel electrophoresis analysis of soluble proteins from RHA1 cells before (A) and after (B) incubation in the presence of biphenyl for 16 h. Proteins were separated in an isoelectric point (pI) gradient from 4 to 7 and then in a 12% polyacrylamide gel, after which they were stained with Coomassie brilliant blue R250. The area of the pI gradient from 5.1 to 5.5 and of molecular mass from 30.0 to 66.0 kDa is shown. The pI and molecular mass scales are indicated at the top and left of each panel, respectively.
Disruption of genes encoding RHDO large subunits.
To examine whether the etbA and ebdA genes are really involved in biphenyl/PCB degradation, etbA1 and ebdA1 were independently disrupted by insertional inactivation using homologous recombination as described in Materials and Methods. The bphA1 gene, which was proved to be involved in biphenyl/PCB degradation in RHA1 (13, 16), was also disrupted by the same method. The etbA1, ebdA1, and bphA1 insertion mutants were designated HDT1, HDB1, and HDA1, respectively.
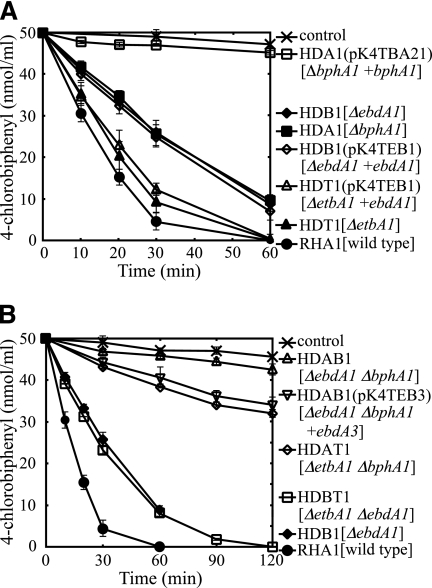
The degradation activity toward 4-CB of each insertion mutant was investigated by GC-MS. Autoclaved cells were used as a negative control. The degradation activities of HDB1 and HDA1 were diminished, and the reduction of 4-CB by either of these insertion mutants was half that by RHA1 after 30 min (Fig. 3A). The activity of HDT1 was only slightly lower than that of RHA1 (Fig. 3A). Taking into account the polar effect exerted on the activities of genes downstream from the inactivated gene, these results suggested that the etbA1, ebdA1, and bphA1 genes and/or the genes downstream from them are involved in biphenyl/PCB degradation.
FIG. 3.
4-Chlorobiphenyl degradation activities of RHA1 and RHDO large-subunit mutants. The results for single-insertion mutants (A) and double-insertion mutants (B) are presented. The remaining amount of 4-chlorobiphenyl was determined by gas chromatography-mass spectrometry. The degradation activities of the derivatives harboring plasmids containing wild-type genes are also shown. Each value is the mean of triplicate independent experiments, with the standard deviation indicated by error bars. Symbols are explained to the right of the panels. Plasmids are indicated in parentheses, and the mutation genotype of a strain and the wild-type gene in a plasmid are indicated in square brackets.
To examine whether the reduction of 4-CB degradation activity was caused by the inactivated gene, a wild-type gene was introduced into the mutant to complement each mutation. A plasmid, pK4TEB1, containing ebdA1 and its promoter region was introduced into the etbA1 mutant, HDT1, and the ebdA1 mutant, HDB1, by electroporation. pK4TBA21, containing bphA1 and its promoter region, was introduced into the bphA1 mutant, HDA1. The degradation activity of each recombinant was then examined. However, none of the 4-CB degradation activities of the insertion mutants were restored (Fig. 3A). In the case of HDA1, the introduction of pK4TBA21 resulted in a further reduction of degradation activity. Excluding the case of HDA1, these results suggested that the expression of RHDO subunit genes located downstream from etbA1 and ebdA1 was suppressed by a polar effect. In the case of HDA1, containing the bphA1 plasmid, the impaired high-level expression of a terminal dioxygenase subunit appears to have resulted in the formation of an inactive heterologous complex between BphA1 and EtbA2/EbdA2.
Double disruption of three RHDO large-subunit genes.
Because the insertion mutants mentioned above did not provide sufficient evidence that RHDO terminal oxygenase component genes play a role in biphenyl/PCB degradation in RHA1, we performed double-disruption experiments. The double-insertion etbA1 bphA1 and ebdA1 bphA1 mutants were constructed by using the bphA1 mutant, HDA1, as the parent strain and were designated HDAT1 and HDAB1, respectively. The double-insertion etbA1 ebdA1 mutant, designated HDBT1, was constructed from the ebdA1 mutant, HDB1.
After examining the ability of the single-disruption mutants to degrade 4-CB, we investigated the 4-CB degradation activity of each double-insertion mutant. The degradation activity of HDAT1 was lower than that of its parent strain, HDA1, and was one-quarter that of RHA1 (Fig. 3B). This degradation activity was estimated to depend solely on the ebdA1A2-encoded terminal oxygenase component (EbdA1A2 component), suggesting that the EbdA1A2 component plays a part in biphenyl/PCB degradation in RHA1. Because HDAT1 was constructed with the etbA1 inactivation of HDA1, the reduction of degradation activity appears to have originated from the inactivation of etbA1. Thus, the terminal oxygenase component encoded by etbA1A2 (EtbA1A2 component) appears to play an important role in biphenyl/PCB degradation in RHA1. On the other hand, the 4-CB degradation activity of HDAB1 was approximately equal to that of the negative control, irrespective of the presence of intact etbA1A2 (Fig. 3B). For HDAB1, it was suggested that the expression of both bphA3 and ebdA3 was inhibited by the polar effect caused by the respective insertions into bphA1 and ebdA1, respectively. Because of the absence of the electron transfer component, the EtbA1A2 component, which is intact in HDAB1, appears to have lost its activity. This notion was supported by the introduction of plasmid pK4TEB3, containing ebdA3, into HDAB1. The degradation activity of HDAB1 was restored by the introduction of pK4TEB3 to a level approximately equal to that of HDAT1 (Fig. 3B). These results again indicated that the EtbA1A2 component is involved in biphenyl/PCB degradation in RHA1. For HDAB1, we conjectured that not only the bphA3 gene but also the adjacent bphA4 gene is inhibited by the polar effect. However, the introduction of the sole ebdA3 gene restored the activity of HDAB1. These results suggested that another ferredoxin reductase gene, probably etbA4, plays a role in biphenyl/PCB degradation.
The degradation activity of HDBT1 was estimated to depend solely on bphA1A2, which confirmed that the bphA1A2-encoded terminal oxygenase component is functional in biphenyl/PCB degradation in RHA1. HDBT1 exhibited almost the same activity as its parent strain, HDB1 (Fig. 3B), although intact etbA1A2 existed in HDB1. These results may suggest that the sole electron transfer components in HDB1 encoded by bphA3 and bphA4 are not sufficient to account for the obvious activity of the etbA1A2-encoded dioxygenase.
Disruption of two RHDO ferredoxin genes and a ferredoxin reductase gene.
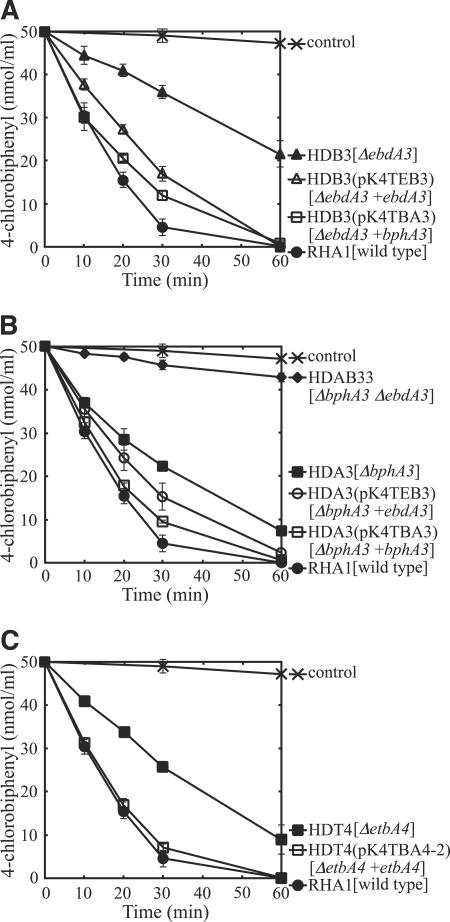
As previously mentioned, ebdA3 and etbA4, which encode a ferredoxin and a ferredoxin reductase, respectively, are thought to be involved in biphenyl/PCB degradation in RHA1. To confirm this involvement, we disrupted these genes by the same method used for the disruption of the RHDO large subunits. The bphA3 gene was also disrupted by the same method. The ebdA3, etbA4, and bphA3 insertion mutants were designated HDB3, HDT4, and HDA3, respectively. We also made repeated attempts to construct a corresponding insertion mutant of bphA4, but these failed.
The ability of each insertion mutant to degrade 4-CB was investigated by GC-MS. The degradation activities of the ferredoxin gene mutants were diminished, with HDB3 and HDA3 realizing one-third and one-half of the 4-CB reduction achieved by RHA1, respectively (Fig. 4). To examine whether the reduction of 4-CB degradation activity was caused by the inactivated gene, a wild-type gene was inserted next to the bphA1 promoter in vector pK4tsr, which was introduced into the mutant to complement each mutation. Plasmid pK4TEB3, containing ebdA3, and pK4TBA3, containing bphA3, were independently introduced into HDB3 and HDA3, respectively, by electroporation. The 4-CB degradation activities of both HDB3 and HDA3 were restored by the introduction of either ebdA3 or bphA3 (Fig. 4A and B). These results indicated that both ferredoxin genes, bphA3 and ebdA3, are involved in biphenyl/PCB degradation. A double-insertion ebdA3 bphA3 mutant was also constructed by using HDA3 as the parent strain and was designated HDAB33. The 4-CB degradation activity of HDAB33 was approximately equal to that of the negative control (Fig. 4B), suggesting that EbdA3 and BphA3 are the only dominant ferredoxin components involved in biphenyl/PCB degradation in RHA1.
FIG. 4.
4-Chlorobiphenyl degradation activities of mutant strains with insertion mutations in RHDO electron transfer components. The results for HDB3 (A), HDA3 (B), and HDT4 (C), with insertion mutations in ebdA3, bphA3, and etbA4, respectively, are presented. The remaining amount of 4-chlorobiphenyl was determined by gas chromatography-mass spectrometry. The degradation activities of the strains harboring plasmids for complementation are also shown. Each value is the mean of triplicate independent experiments, with the standard deviation indicated by error bars. Symbols are explained to the right of the panels. Plasmids are indicated in parentheses, and the mutation genotype of a strain and the wild-type gene in a plasmid are indicated in square brackets.
The degradation activity of the ferredoxin reductase mutant HDT4 was diminished to about half that of RHA1 (Fig. 4C). To complement the etbA4 gene deficiency, pK4TBA4-2, containing etbA4 and its promoter region, was introduced into HDT4 by electroporation. The 4-CB degradation activity of HDT4 was restored by introduction of the etbA4 gene (Fig. 4C). These results indicated that a ferredoxin reductase gene of RHDO, etbA4, is involved in biphenyl/PCB degradation in RHA1.
Transformation of PCB congeners by insertion mutants.
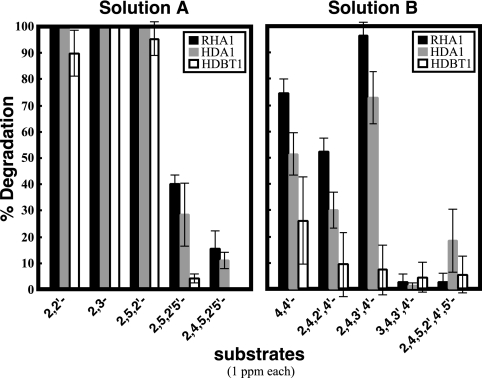
It is known that the large subunits of terminal oxygenase components are major determinants of the substrate preference of the RHDO. In consideration of the low sequence similarity between EtbA1A2/EbdA1A2 and BphA1A2, it was expected that the substrate preferences would differ between EtbA/EbdA and BphA dioxygenase species. Therefore, the substrate preferences of EtbA/EbdA and BphA for PCB congeners were investigated by using the insertion mutants. The bphA1 mutant, HDA1, and the etbA1 ebdA1 mutant, HDBT1, were employed as EtbA/EbdA- and BphA-expressing strains, respectively. A mixture of five ortho-substituted PCB congeners that consisted of 2,2′-, 2,3-, 2,5,2′-, 2,5,2′,5′-, and 2,4,5,2′,5′-chlorobiphenyl (solution A) and a mixture of five para-substituted PCB congeners that consisted of 4,4′-, 2,4,2′,4′-, 2,4,3′,4′-, 3,4,3′,4′-, and 2,4,5,2′,4′,5′-chlorobiphenyl (solution B) were used as substrates. The results are shown in Fig. 5. Although the degradation activity of HDA1 was lower than that of RHA1, the substrate preference of EtbA/EbdA-expressing HDA1 was almost identical to that of RHA1. On the other hand, BphA-expressing HDBT1 showed extremely low degradation activities toward 2,4,2′,4′-, 2,4,3′,4′-, 2,5,2′,5′-, and 2,4,5,2′,5′-chlorobiphenyls. The results for HDBT1 were almost correspondent with those obtained by using Rhodococcus erythropolis IAM1399 expressing bphA1A2A3A4 (16). These results indicated that EtbA/EbdA and BphA have different substrate preferences for PCB congeners and suggested that EtbA/EbdA plays an important role in the degradation of highly chlorinated biphenyls. BphA appears to play a role in the degradation of PCBs to some extent, because HDBT1 showed good activities toward 2,2′-, 2,3-, and 2,5,2′-chlorobiphenyls in addition to 4-CB (Fig. 3 shows the data for 4-CB).
FIG. 5.
Degradation of PCB congeners by RHA1 (black bars), by the bphA1 mutant, HDA1 (gray bars), and by the etbA1 ebdA1 double knockout, HDBT1 (white bars). Solution A consisted of five mainly ortho-substituted PCB congeners, 2,2′-dichlorobiphenyl, 2,3-dichlorobiphenyl, 2,5,2′-trichlorobiphenyl, 2,5,2′,5′-tetrachlorobiphenyl, and 2,4,5,2′,5′-pentachlorobiphenyl. Solution B consisted of five mainly para-substituted PCB congeners, 4,4′-dichlorobiphenyl, 2,4,2′,4′-tetrachlorobiphenyl, 2,4,3′,4′-tetrachlorobiphenyl, 3,4,3′,4′-tetrachlorobiphenyl, and 2,4,5,2′,4′,5′-hexachlorobiphenyl. Each value is the mean of triplicate independent experiments, with the standard deviation indicated by error bars.
Multiple isozyme genes for biphenyl/PCB degradation in Rhodococcus strains have been reported (9, 11, 18). However, the potential involvement of these isozyme genes in degradation has not been investigated. To our knowledge, this is the first report to demonstrate the cooperative involvement of multiple RHDO isozymes encoded by the bphA, etbA, and ebdA genes in a single PCB-degrading bacterium. These genes were indicated to be under the control of the bphST regulatory system, which has a broad substrate spectrum for transcription induction (18, 19). Thus, these isozymes might be simultaneously involved in the catabolism of substrates other than biphenyl.
Acknowledgments
We thank René De Mot for the kind gift of plasmid pFAJ2574. We are grateful to R. van der Geize for his helpful suggestions regarding the insertional inactivation of rhodococcal genes.
This study was supported in part by a grant-in-aid for the study of hazardous chemicals from the Ministry of Agriculture, Forestry, and Fisheries of Japan (HC-05-2322-1).
REFERENCES
- 1.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, L. G. Seidman, J. A. Smith, and K. Struhl. 1990. Current protocols in molecular biology. John Wiley and Sons, Inc., New York, N.Y.
- 2.Bedard, D. L., M. L. Haberl, R. J. May, and M. J. Brennan. 1987. Evidence for novel mechanisms of polychlorinated biphenyl metabolism in Alcaligenes eutrophus H850. Appl. Environ. Microbiol. 53:1103-1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.De Mot, R., I. Nagy, A. De Schrijver, P. Pattanapipitpaisal, G. Schoofs, and J. Vanderleyden. 1997. Structural analysis of the 6 kb cryptic plasmid pFAJ2600 from Rhodococcus erythropolis NI86/21 and construction of Escherichia coli-Rhodococcus shuttle vectors. Microbiology 143:3137-3147. [DOI] [PubMed] [Google Scholar]
- 4.Furukawa, K., and F. Matsumura. 1976. Microbial metabolism of polychlorinated biphenyls. Studies on the relative degradability of polychlorinated biphenyl components by Alcaligenes sp. J. Agric. Food Chem. 24:251-256. [DOI] [PubMed] [Google Scholar]
- 5.Furukawa, K., N. Tomizuka, and A. Kamibayashi. 1979. Effect of chlorine substitution on the bacterial metabolism of various polychlorinated biphenyls. Appl. Environ. Microbiol. 38:301-310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hashimoto, Y., M. Nishiyama, F. Yu, I. Watanabe, S. Horinouchi, and T. Beppu. 1992. Development of a host-vector system in a Rhodococcus strain and its use for expression of the cloned nitrile hydratase gene cluster. J. Gen. Microbiol. 138:1003-1010. [DOI] [PubMed] [Google Scholar]
- 7.Kimbara, K., T. Hashimoto, M. Fukuda, T. Koana, M. Takagi, M. Oishi, and K. Yano. 1989. Cloning and sequencing of two tandem genes involved in degradation of 2,3-dihydroxybiphenyl to benzoic acid in the polychlorinated biphenyl-degrading soil bacterium Pseudomonas sp. strain KKS102. J. Bacteriol. 171:2740-2747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kitagawa, W., K. Miyauchi, E. Masai, and M. Fukuda. 2001. Cloning and characterization of benzoate catabolic genes in the gram-positive polychlorinated biphenyl degrader Rhodococcus sp. strain RHA1. J. Bacteriol. 183:6598-6606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kitagawa, W., A. Suzuki, T. Hoaki, E. Masai, and M. Fukuda. 2001. Multiplicity of aromatic ring hydroxylation dioxygenase genes in a strong PCB degrader, Rhodococcus sp. strain RHA1 demonstrated by denaturing gradient gel electrophoresis. Biosci. Biotechnol. Biochem. 65:1907-1911. [DOI] [PubMed] [Google Scholar]
- 10.Kohler, H. P., D. Kohler-Staub, and D. D. Focht. 1988. Cometabolism of polychlorinated biphenyls: enhanced transformation of Aroclor 1254 by growing bacterial cells. Appl. Environ. Microbiol. 54:1940-1945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Maeda, M., S. Y. Chung, E. Song, and T. Kudo. 1995. Multiple genes encoding 2,3-dihydroxybiphenyl 1,2-dioxygenase in the gram-positive polychlorinated biphenyl-degrading bacterium Rhodococcus erythropolis TA421, isolated from a termite ecosystem. Appl. Environ. Microbiol. 61:549-555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Masai, E., K. Sugiyama, N. Iwashita, S. Shimizu, J. E. Hauschild, T. Hatta, K. Kimbara, K. Yano, and M. Fukuda. 1997. The bphDEF meta-cleavage pathway genes involved in biphenyl/polychlorinated biphenyl degradation are located on a linear plasmid and separated from the initial bphACB genes in Rhodococcus sp. strain RHA1. Gene 187:141-149. [DOI] [PubMed] [Google Scholar]
- 13.Masai, E., A. Yamada, J. M. Healy, T. Hatta, K. Kimbara, M. Fukuda, and K. Yano. 1995. Characterization of biphenyl catabolic genes of gram-positive polychlorinated biphenyl degrader Rhodococcus sp. strain RHA1. Appl. Environ. Microbiol. 61:2079-2085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mondello, F. J. 1989. Cloning and expression in Escherichia coli of Pseudomonas strain LB400 genes encoding polychlorinated biphenyl degradation. J. Bacteriol. 171:1725-1732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sakai, M., K. Miyauchi, N. Kato, E. Masai, and M. Fukuda. 2003. 2-Hydroxypenta-2,4-dienoate metabolic pathway genes in a strong polychlorinated biphenyl degrader, Rhodococcus sp. strain RHA1. Appl. Environ. Microbiol. 69:427-433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Seto, M., E. Masai, M. Ida, T. Hatta, K. Kimbara, M. Fukuda, and K. Yano. 1995. Multiple polychlorinated biphenyl transformation systems in the gram-positive bacterium Rhodococcus sp. strain RHA1. Appl. Environ. Microbiol. 61:4510-4513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Seto, M., K. Kimbara, M. Simura, T. Hatta, M. Fukuda, and K. Yano. 1995. A novel transformation of polychlorinated biphenyls by Rhodococcus sp. strain RHA1. Appl. Environ. Microbiol. 61:3353-3358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Takeda, H., N. Hara, M. Sakai, A. Yamada, K. Miyauchi, E. Masai, and M. Fukuda. 2004. Biphenyl-inducible promoters in a polychlorinated biphenyl-degrading bacterium, Rhodococcus sp. RHA1. Biosci. Biotechnol. Biochem. 68:1249-1258. [DOI] [PubMed] [Google Scholar]
- 19.Takeda, H., A. Yamada, K. Miyauchi, E. Masai, and M. Fukuda. 2004. Characterization of transcriptional regulatory genes for biphenyl degradation in Rhodococcus sp. strain RHA1. J. Bacteriol. 186:2134-2146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yamada, A., H. Kishi, K. Sugiyama, T. Hatta, K. Nakamura, E. Masai, and M. Fukuda. 1998. Two nearly identical aromatic compound hydrolase genes in a strong polychlorinated biphenyl degrader, Rhodococcus sp. strain RHA1. Appl. Environ. Microbiol. 64:2006-2012. [DOI] [PMC free article] [PubMed] [Google Scholar]