Abstract
We determined whether a recently developed method to isolate specific small-subunit (SSU) rRNAs can be used in 13C-labeling studies to directly link community structure and function in natural ecosystems. Replicate North Sea sediment cores were incubated at the in situ temperature following addition of 13C-labeled acetate, propionate, amino acids, or glucose. Eukaryotic and bacterial SSU rRNAs were separated from total RNA by means of biotin-labeled oligonucleotide probes and streptavidin-coated paramagnetic beads, and the 13C content of the isolated rRNA was determined by elemental analysis-isotope ratio mass spectrometry. The SSU rRNA yield with the bead-capture protocol was improved by using helper probes. Incorporation of label into bacterial SSU rRNA was detectable after 2 h of incubation. The labeling was always much greater in bacterial SSU rRNA than in eukaryotic SSU rRNA, suggesting that bacteria were the main consumers of the 13C-labeled compounds. Similar results were obtained with the 13C-labeled polar-lipid-derived fatty acid (PLFA) approach, except that more label was detected in bacterial PLFA than in bacterial SSU rRNA. This may be attributable to the generally slow growth of sediment microbial populations, which results in low ribosome synthesis rates and relatively few ribosomes per cell. We discuss possible ways to improve the probe-capture protocol and the sensitivity of the 13C analysis of the captured SSU rRNA.
Coastal sediments are often highly active, and much of the carbon processing in them is carried out by the microbial community (18), which can be very diverse (14, 37, 57). As in most natural ecosystems, only a small fraction of the microbial species detected in coastal sediments have been grown in pure culture (for a review see reference 33), so it is often not known which organisms are responsible for which steps in carbon cycling. Several cultivation-independent methods directly linking biogeochemical processes and the organisms carrying them out have been developed. Boschker et al. (13) labeled sediments with 13C-substrates and monitored label incorporation into lipid biomarkers. This approach has been used to directly link function with specific subsets of the microbial community (11, 17, 32, 53) and to study the role of microbes in food webs (48). However, the phylogenetic resolution possible with lipids, such as polar-lipid-derived fatty acids (PLFA), is limited (12, 63).
Small-subunit (SSU) rRNA or rRNA gene sequences are widely used to identify microorganisms in natural systems (16, 43, 62), and they can provide much greater phylogenetic resolution than lipids. Several methods that combine the advantages of molecular community analysis and labeling with 13C-substrates have been developed. Radajewski et al. (55) introduced stable isotope probing (SIP), which is based on separation of highly labeled, heavy [13C]DNA from lighter unlabeled material by ultracentrifugation in a density gradient. The DNA isolated from the heavy fraction can be analyzed subsequently by a range of molecular techniques in order to identify the 13C-labeled, active microorganisms. An rRNA-based SIP method has also been developed, which allows more sensitive analysis, as the rRNA is labeled more quickly than DNA (47). However, a disadvantage of the SIP method is that the rRNA or DNA must incorporate >50% of the 13C to be separated from unlabeled material. This is especially a problem in natural ecosystems with low levels of organic matter. Although improvements have been made (30, 42), large amounts of label or long incubation times are still needed, which may stimulate fast-growing organisms that are not active under natural conditions.
Another approach is based on isolation of specific rRNAs with a magnetic bead probe-capture protocol, followed by isotope ratio mass spectroscopy (IRMS) analysis of the isolated material (44, 52). Like the 13C-labeled lipid method, IRMS allows detection of incorporation of label into rRNA at very low levels. Labeled substrates can therefore be added at concentrations that are close to natural concentrations, avoiding possible artifacts (12). So far, the probe-capture 13C-rRNA method has been used only for natural-abundance 13C work with pure cultures (44, 52); studies of symbionts of cold seep and hydrothermal vent mussels, in which a strong isotopic signature from methane is expected, are in progress (7).
In the study reported here, coastal sediment was labeled with several 13C-substrates, and incorporation of label into bacterial and eukaryotic rRNA and PLFA biomarkers was examined. In addition, we improved the efficiency of probe capture by including unlabeled helper probes (29). Both the lipid and rRNA methods indicated that bacteria were the main consumers of the organic substrates tested; very little label could be traced to eukaryotic biomarker SSU rRNA or PLFA. However, the relative and absolute levels of labeling were different for the two types of biomarkers. Below we discuss possible causes for these differences and possible ways to improve the probe-capture protocol and 13C analysis of the captured SSU rRNA.
MATERIALS AND METHODS
Sampling and 13C labeling.
Small sediment cores were obtained from an intertidal mud flat just outside the Ritthem salt marsh in the Westerscheldt estuary (The Netherlands) on 14 April 2003. The sediment consisted of fine sand with 22% silt, and the salinity in this part of the estuary is approximately 29‰. Cores were collected using 25-ml disposable syringes with the tips cut off. The top 6 cm of the sediment was sampled, and samples were transferred directly to the laboratory, where they were incubated in the dark at the in situ temperature (14°C).
Labeling was started by five injections of 9 μl of 13C-labeled substrate into the top 5 cm of the sediment by the line injection method (35). This distributed the label evenly throughout the sediment core with as little disturbance as possible. The substrates added were uniformly labeled [13C]acetate (200 mM in Milli-Q), [13C]glucose (100 mM), and 13C-amino acids (100 mM), as well as [13C]propionate (200 mM) labeled at the methyl position (for all substrates, >98% 13C; Cambridge Isotope Laboratories, United States). The concentrations of the added substrates were varied to keep the final concentration, in terms of carbon, more or less constant (900 to 1,350 μmol C/liter sediment). Sediments labeled with [13C]acetate were incubated in a time series between 2 and 48 h, and sediments labeled with the other three substrates were incubated for 6 h. All treatments were performed in quadruplicate, and two cores each were used for PLFA analysis and rRNA extraction. Sediment cores were frozen immediately after incubation at −20°C for PLFA analysis and at −80°C for rRNA analysis. PLFA samples were freeze-dried before analysis. Unlabeled control cores were also processed.
RNA isolation and electrophoresis.
All solutions were prepared with diethylpyrocarbonate-treated distilled, deionized water (RNase-free distilled, deionized H2O). RNA was isolated from approximately 30 g of sediment per sample. Cells were lysed by bead beating with an MSK-Zellhomogenisator (B. Braun Biotech International, Melsungen, Germany), using a 70-ml stainless steel chamber and 40 s of bead beating at 4,000 reciprocations per min. RNA was then extracted and purified by phenol-chloroform extraction and isopropanol precipitation (46, 61). The final RNA pellet was resuspended in 400 μl RNase-free distilled, deionized H2O. Aliquots of each RNA sample were separated by electrophoresis on two-phase (3.3%/10%) polyacrylamide gels (3). Nucleic acid bands were visualized by ethidium bromide staining.
Magnetic bead capture hybridization.
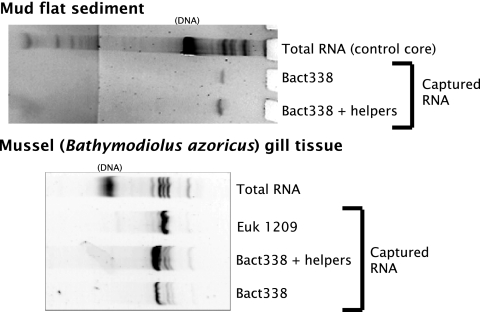
Oligonucleotide probes and helper probes (Table 1) were purchased from ThermoHybaid (Ashford, Middlesex, England). Magnetic bead capture hybridization was performed essentially as previously described (44), except that 7.5× SSC was used for the posthybridization bead washes to increase the rRNA yield (6) (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate). To further increase the yield, unlabeled 21-mer helper probes (29) complementary to the consensus sequences upstream and downstream of the Bact338 and Euk1369 probe target sites were designed, using the sequence database analysis program ARB (41). When tested, helper probes increased the capture of specific rRNAs severalfold (Fig. 1), presumably by “melting out” the secondary structure around the probe target site.
TABLE 1.
Oligonucleotide probes and helper probes used in this study
| Probea | Abbreviation | Target group | Sequence (5′-3′)b | Reference |
|---|---|---|---|---|
| S-D-Bact-0338-a-A-18 | Bact338 | Most bacteriac | GCT GCC TCC CGT AGG AGT | 4 |
| S-D-Euca-1379-a-A-16 | Euk1379 | Most eukaryotes | TAC AAA GGG CAG GGA C | 34 |
| S-D-Euca-1209-a-A-16 | Euk1209 | Most eukaryotes | GGG CAT CAC AGA CCT G | 40 |
| Bact338_up_help | Bacteria | CTG GNC CGT GTC TCA GTN CCA | This study | |
| Bact338_down_help | Bacteria | CAT TGY SCA ANA TTC CCC ACT | This study | |
| Euk1379_up_help | Eukaryotes | GTA ATC AAC GCR AGC TGA TGA | This study | |
| Euk1379_down_help | Eukaryotes | GTA GKA GCG ACG GGC GGT GTG | This study |
The two capture probes were named in accordance with the suggestions of Alm et al. (2).
S = G or C; R = A or G; Y = C or T; K = G or T; N = G, A, T, or C.
Bact338 targets (many) chloroplasts and other plastids but not mitochondria.
FIG. 1.
Examples of helper probes used for bead capture hybridization. The unlabeled upstream and downstream Bact338 helper probes (Table 1) were used at a 1:1:1 ratio with the biotin-labeled Bact338 capture probe. Note that the gill samples included both prokaryotic and eukaryotic rRNAs; Bathymodiolus azoricus gills harbor symbiotic bacteria (7).
13C analysis of captured rRNA.
Captured rRNA was analyzed with an elemental analyzer (EA) (Carlo Erba, Italy) coupled to an isotope ratio mass spectrometer (Delta Plus; Thermo Finnigan, Germany). The IRMS was tuned for maximum sensitivity, and the source was cleaned shortly before analysis. The sensitivity was increased further by measurement with helium dilution turned off on the Conflo interface (Thermo Finnigan, Germany) between the EA and the IRMS. The captured rRNA was transferred to silver cups, and blank cups with either 100 μl distilled, deionized H2O or nothing added were also analyzed. These blank cups contained approximately 0.3 μg C with a δ13C ratio of −26.8‰. The measured δ13C ratios of the rRNA samples were corrected for blanks using a simple mass balance equation. This allowed us to analyze as little as ∼0.5 μg C of rRNA after blank correction, and samples usually contained between 1 and 2 μg C of rRNA.
PLFA extraction and analysis.
Freeze-dried sediments were extracted and PLFA were analyzed as described by Boschker (10). Briefly, lipids were extracted in chloroform-methanol-water using a modified Bligh-Dyer method and were fractionated on silicic acid into different polarity classes. The most polar fraction, containing the PLFA, was derivatized to obtain fatty acid methyl esters (FAME). The concentration and carbon isotopic composition of individual FAME were determined with a gas chromatograph-combustion interface-isotope ratio mass spectrometer (GC-c-IRMS) consisting of an HP G1530 gas chromatograph (Hewlett-Packard, United States) connected to a Delta-plus IRMS via a type III combustion interface from Thermo Finnigan (Germany). Internal (12:0 and 19:0) and external FAME reference mixtures were used to check the accuracy of the isotopic ratios determined by the GC-c-IRMS. Stable carbon isotope ratios for individual PLFA were calculated from FAME data by correcting for the one carbon atom in the methyl group that was added during derivatization. Specific biomarkers used for Bacteria and Eukarya are listed in Table 2.
TABLE 2.
Specific PLFA biomarkers used in this study
| Group | PLFA biomarkers |
|---|---|
| Bacteria | i14:0, i15:0, a15:0, i16:0, i17:1ω7, 10Me16:0, a17:1ω7, i17:0, a17:0, cy17:0, 18:1ω7c, cy19:0 |
| Eukaryotes | 20:4ω6, 20:5ω3a |
Minor amounts of other eukaryotic, polyunsaturated PLFA (16:PUFA, 18:2ω6) were also detected, but these compounds were not completely separated from PLFA found in bacteria and therefore were not used.
Calculations.
All primary stable carbon isotope data below are in the delta notation (δ13C expressed in ‰) relative to the Vienna PDB. As this was a 13C labeling study, we expressed most of our results as either the pool average difference in δ13C ratio between a labeled sample and the unlabeled controls (Δδ13C) or as the percentage of the label recovered based on excess 13C data (10).
RESULTS
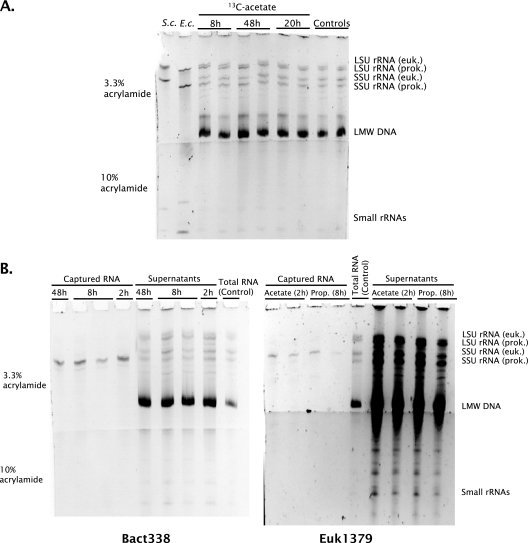
Gels prepared from the total RNA extracts showed that comparable amounts of eukaryotic and bacterial long-subunit and SSU rRNAs were extracted (Fig. 2A), as expected for a coastal sediment covered by benthic diatoms. Figure 2B shows the selectivity of the probe-capture method, and the results indicate that bacterial SSU rRNA in the mixture of extracted RNAs could be specifically isolated. Similar selectivity was achieved for the capture of eukaryotic SSU rRNA. The bacterial SSU rRNA capture efficiency was estimated to be 16% ± 5% for a total of 11 ± 1 μg bacterial SSU rRNA/g (dry weight) (n = 16), based on the intensities of the bands in Fig. 2A and B. Eukaryotic SSU rRNA could not be quantified in this way because the band intensities on the gels used to check rRNA quality were too strong compared to the standards and the remaining material was needed for isotope analysis.
FIG. 2.
(A) Examples of sediment-extracted total RNA. The RNA in each of the sample lanes represents approximately 7.5 mg of sediment, out of a total of approximately 30 g. The pure-culture reference RNAs were Saccharomyces cerevisiae RNA (S.c.) and Escherichia coli RNA (E.c.) (approximately 200 ng each). (B) Examples of magnetic bead capture hybridization with the Bact338 and Euk1379 probes. LSU, long subunit; euk., eukaryote; prok., prokaryote; LMW, low molecular weight.
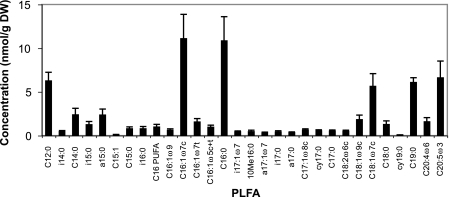
Concentrations of individual PLFA provide an indication of the community biomass and composition in sediments. The total PLFA concentration found in the sediment studied was 55 ± 16 nmol/g (dry weight) (n = 16). The PLFA detected were characteristic of intertidal mud flat sediments, and there were substantial amounts of both bacterium- and eukaryote-specific PLFA (Fig. 3 and Table 2). Eukaryotic PLFA were probably derived primarily from benthic diatoms, which were clearly visible on the sediment surface during sampling, as they were dominated by the typical diatom PLFA 20:5ω3. The most abundant bacterial PLFA detected were i15:0, a15:0, and 18:1ω7c. Bacterial PLFA accounted for 23% ± 1% of the total PLFA, and eukaryotic PLFA accounted for 18% ± 3%; the remainder were common PLFA, such as 16:1ω7c and 16:0, which are found in both bacteria and eukaryotes.
FIG. 3.
PLFA concentrations. The values are averages ± standard deviations for all samples (n = 16). DW, dry weight; PUFA, polyunsaturated fatty acid.
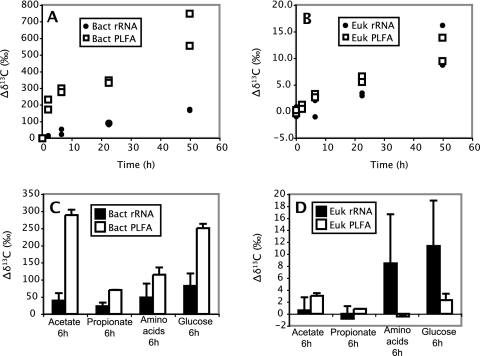
In the [13C]acetate time series experiment, both rRNA and PLFA were significantly labeled after 2 h of incubation (Fig. 4A and B). The PLFA data shown in Fig. 4 are pool-averaged data for bacterium- or eukaryote-specific PLFA, which allow direct comparison with the rRNA results. Bacterial rRNA and PLFA were both labeled to a much greater extent than eukaryotic biomarkers, suggesting that bacteria were the principal consumers of the label. There was, however, a striking difference between the levels of labeling in bacterial rRNA and bacterial PLFA. When the data were expressed as Δδ13C ratios, bacterial PLFA were labeled between 4 and 13 times more than bacterial rRNA. The bacterial PLFA were also labeled substantially faster than the bacterial rRNA, especially during the first 6 h of the [13C]acetate time series. The differences between eukaryotic rRNA and eukaryotic PLFA were smaller. Labeling of eukaryotic rRNA appeared to lag behind labeling of eukaryotic PLFA, but by the end of the time course the levels of labeling for the two eukaryotic biomarkers were similar.
FIG. 4.
Labeling of rRNA and pool average PLFA expressed as Δδ13C values. (A and B) Results of the [13C]acetate time series for bacterial (A) and eukaryotic (B) markers. (C and D) Comparison of different 13C-labeled substrates. Bact, bacterial; Euk, eukaryotic.
The timing of the labeling differed among the bacterial PLFA, as shown in Fig. 5 for two of the most abundant biomarkers. The PLFA 18:1ω7c (Fig. 5), 16:1ω7c, and 16:0 were labeled very quickly during the first 6 h, and this was followed by slower or even decreased labeling until 22 h and a subsequent increase from 22 to 48 h. As 18:1ω7c is one of the most abundant bacterium-specific PLFA in the sediment studied (Fig. 3), this pattern is also evident in the pool-averaged labeling data shown in Fig. 4. Other PLFA, such as a15:0, were labeled more gradually, and the labeling rate decreased at the end of incubation (Fig. 5).
FIG. 5.
13C incorporation into two main bacterial PLFA, as determined in the [13C]acetate time series experiment. DW, dry weight.
The substrate comparison experiment gave results similar to those of the acetate time series experiment, with most of the label in bacterial biomarkers and greater labeling of bacterial PLFA than of bacterial rRNA (Fig. 4C and D). Differences between the two bacterial biomarkers were especially pronounced for [13C]acetate (ninefold-higher Δδ for PLFA). For the other substrates, the difference was approximately threefold, and there was substantial variation between replicates (range, 1.8- to 5.2-fold). The labeling levels were also different for bacterial PLFA, with much lower Δδ13C ratios with 13C-amino acids and propionate, although the latter can be explained to some extent by the use of propionate that was labeled only at the methyl carbon atom. Eukaryotic biomarkers showed no clear pattern for different substrates, and the levels of labeling were generally low (Fig. 4C and D). Labeling of eukaryotic rRNA with [13C]acetate and [13C]propionate could not be detected, whereas eukaryotic PLFA were not labeled with 13C-amino acids. With 13C-amino acids and [13C]glucose, more label was incorporated into eukaryotic rRNA than into eukaryotic PLFA, although the levels of labeling were low and highly variable (especially for the rRNA). In general, the levels of labeling were more variable for rRNA than for PLFA.
The percentage of label recovered in PLFA was calculated from the amounts of label added, the measured PLFA concentrations, and the levels of labeling, using the equations described by Boschker (10). The level of label recovery in bacterial PLFA ranged from 0.02% for [13C]propionate to 0.08% for [13C]acetate after 6 h of incubation (Fig. 6B). A maximum of 0.2% label was found in bacterial PLFA after 48 h of incubation with [13C]acetate (Fig. 6A). As indicated above, in all incubations the most label was found in bacterium-specific marker PLFA (30 to 55%), and little label was detected in eukaryotic PLFA (<1%). The percentage of label recovered was also estimated for bacterial rRNA, based on the levels of labeling shown in Fig. 4 and a concentration of 3.8 ± 0.4 μg C/g (dry weight) estimated from the rRNA gels calibrated with known amounts of an RNA sizing ladder (Fig. 2). These calculations indicated that the levels of label recovery in bacterial rRNA were comparable for the different substrates after 6 h of incubation (Fig. 6B). Between 1.4 and 2.3 times more label was recovered in bacterial PLFA than in bacterial rRNA with [13C]propionate, 13C-amino acids, and glucose, and approximately 5 times more label was recovered with [13C]acetate. In general, the [13C]acetate time series also showed that there was a higher level of label recovery in bacterial PLFA than in bacterial rRNA (between 1.4 and 8.4 times higher in PLFA). The differences between labeling in PLFA and labeling in rRNA were always smaller when the data were expressed as percentages of label recovered than when the data were expressed as Δδ ratios.
FIG. 6.
Percentage of the added label incorporated into bacterial PLFA and bacterial rRNA for the [13C]acetate time series (A) and for the different substrates after 6 h (B). Note the difference the in y axis scales. Bact, bacterial.
DISCUSSION
We found that 13C label incorporation into particular classes of rRNA can be monitored to study carbon utilization in natural microbial communities in sediments. Sufficient rRNA could be extracted and isolated by the magnetic bead capture protocol to allow accurate 13C analysis by EA-IRMS. The distribution of label between bacterial and eukaryotic SSU rRNA suggests that all organic substrates tested were predominantly used by bacteria. The label incorporation by eukaryotes, which were probably predominately benthic diatoms, was limited. Similar patterns were found when the label distribution in specific biomarker PLFA was analyzed. The obvious next step is to use the 13C-labeled rRNA method with probes targeting more specific phylogenetic groups. This should allow us to study sedimentary carbon cycling with much greater phylogenetic resolution than is possible with lipid biomarkers.
With regard to probe specificity, it should be pointed out that the bacterial PLFA and rRNA analyzed may not have been derived from identical populations. For PLFA, some species may not contain particular “domain-specific” lipids, so labeling may be underestimated. For rRNA, there are several known problems with the Bact338 and Euk1379 probes, which were designed more than 10 years ago (4, 34). Euk1379 misses some major eukaryotic groups, but it does target nearly all Bacillariophyta (diatom) SSU rRNAs currently in the GenBank database (not shown). The probe mixture of Baker et al. (5) could be used in future experiments. The major groups known to be missed by Bact338 are the Planctomycetales, Chlamydiae, Verrucomicrobiae, and Aquificales. Depending on the prevalence of nontarget species, their rates of label incorporation, and their substrate preferences, bacterial label incorporation might be either over- or underestimated. Planctomycetales, at least, are often found in marine sediments (28, 56). The probe combination of Daims et al. (20) could be used for most of the missed groups. Bact338 also targets some organellar rRNAs, particularly those of chloroplasts, which of course are found in diatoms.
Although the rRNA and PLFA results were similar in terms of bacterial versus eukaryotic uptake of organic substrates, there were conspicuous differences in the levels of bacterial labeling between the two methods. In general, label incorporation was substantially greater in bacterial PLFA than in bacterial rRNA, especially when the results were expressed as Δδ13C values. As the data are based on 13C/12C ratios, they do not express absolute label incorporation but rather express labeling relative to the carbon pool studied. This makes Δδ13C ratios vulnerable to dilution by unlabeled carbon pools. These pools may include protocol blanks or unlabeled detrital material extracted along with rRNA or PLFA. This is especially true for the rRNA because samples were analyzed by EA-IRMS, which measures bulk carbon. The PLFA data are much less likely to be affected by contaminants because analytes are separated at a high resolution by gas chromatography before combustion for IRMS. However, it seems unlikely that materials other than SSU rRNA passed through the multiple fractionation and wash steps of the probe-capture protocol. Unlabeled detrital rRNA could dilute the 13C signal, but it is generally thought that RNA has a high turnover rate and does not accumulate in sediments (22). MacGregor et al. (44) rigorously tested the rRNA method for protocol blanks and found that for the most part these can be avoided by careful selection of solvents and buffers. In the future we hope to circumvent such problems by analyzing high-performance liquid chromatography (HPLC)-separated nucleic acids (45).
Absolute label recovery for both bacterial biomarkers showed a similar but less pronounced trend: more label was recovered in bacterial PLFA than in bacterial rRNA. The percentages of label recovered represent the SSU rRNA and PLFA contents of the bacterial biomass synthesized from the added 13C label. Based on pure-culture work, one would expect bacterial cell mass to be between 5 and 20% RNA (49, 50). Assuming that about 25% of RNA is SSU rRNA (50) and that new PLFA and rRNA are synthesized at comparable rates, similar amounts of label should have been detected in bacterial SSU rRNA and PLFA (between 1 and 5%) (15). Dilution of the rRNA sample with unlabeled materials, if any, would not affect the percentage of label recovered. However, the amounts are influenced by biomarker extraction efficiency. In our experience, approximately 85% of sediment PLFA is generally recoverable (data not shown). Experiments in which Shewanella putrefaciens cultures were mixed with lake sediment, the total RNA was extracted, and SSU rRNA was measured by slot blot hybridization suggested that rRNA recovery could be in the same range (MacGregor, unpublished observations); however, the added cells were likely larger, less firmly attached, and possibly more easily lysed than native bacteria.
The PLFA content of bacteria is relatively constant, and an average of 0.056 g PLFA C/g biomass C gives very reasonable biomass and total 13C incorporation rates for sedimentary bacteria (48). rRNA content, on the other hand, varies among species (19, 26) and is generally growth rate dependent in both prokaryotes (8, 38, 58, 59) and eukaryotes (36, 64), although at least some bacterial species maintain a ribosomal reserve under starvation conditions (27, 54). A relatively low growth rate and associated low cellular ribosome content might explain the low level of label recovery in bacterial rRNA compared to bacterial PLFA.
The level of label recovery in bacterial PLFA was rather variable among substrates and was especially high in sediments labeled with [13C]acetate, whereas the labeling in bacterial rRNA was more constant. This may reflect differences in growth efficiency or preferential labeling of PLFA with particular substrates. Acetate, or more precisely acetyl coenzyme A, is a direct and the most important fatty acid precursor, so it is not unexpected that a relatively high proportion of the [13C]acetate label was incorporated into PLFA. The different labeling rates for PLFA could be explained by the presence of two communities in the sediment, a fast-growing community containing relatively large amounts of quickly labeled compounds (e.g., 16:1ω7c and 18:1ω7c) and a slowly growing community containing larger amounts of, for instance, a15:0.
rRNA has a more complex biosynthetic pathway than PLFA, and it is composed of both ribose (synthesized from glucose) and pyrimidine and purine bases (synthesized primarily from amino acids) (51). rRNA may therefore be less prone to preferential incorporation, allowing a better relationship between growth rates and labeling patterns and better comparisons among substrates. On the other hand, rRNA labeling may be affected more by cross-feeding and by processes including natural transformation (23, 24) and nucleoside (1) or nucleobase (21) uptake. The significance of these factors in the sediment microbial community and of possible preformed lipids and lipid precursors (9, 25, 65) is not known.
For the intertidal sediment studied, a minimum of 2 to 5 g of sediment was needed to obtain sufficient bacterial SSU rRNA (∼1 μg) for a single analysis. This restricts the method to active, high-biomass samples and more abundant microbial groups until the EA-IRMS sensitivity and/or rRNA capture efficiency can be improved. Helium flow rates of ∼80 ml/min are needed for the EA, whereas the IRMS can take less than 1 ml/min; up to 99% of the CO2 produced by sample combustion in the EA is therefore lost. More efficient transfer could decrease the RNA requirement some 50-fold. Further improvements could result from analyzing rRNA nucleotides by HPLC-IRMS (39) or GC-c-IRMS, which have much better transfer efficiencies and allow direct identification of the compounds being analyzed (45). Furthermore, the HPLC-IRMS interface can be used as a micro-EA-IRMS if very small samples are injected (31, 60). Use of 14C-labeled substrates with detection by scintillation counting or autoradiography could also increase the sensitivity. In this study, we increased the rRNA capture efficiency by using helper probes alongside the capture probe. Spacers between the magnetic beads and the capture probes might help further by limiting steric hindrance. Given the potential for improving the probe-capture method, we expect that it will be possible to study carbon incorporation by major phylogenetic groups in microbial communities with greatly improved resolution and perhaps also to investigate naturally occurring differences in carbon isotope distribution.
Acknowledgments
We thank Peter van Breugel, Yvonne van de Maas, and Marco Houtekamer for assistance with the 13C analysis. Claudia Bergin and Nicole Dubilier (MPI-Bremen) are gratefully acknowledged for providing the Bathymodiolus azoricus data in Fig. 1.
This work was supported by NWO-VIDI grant 864.04.009, by a grant from the Fungal-Bacterial Interactions project of the Vernieuwingsfonds of the Royal Netherlands Academy of Arts and Sciences (KNAW) to H.T.S.B., and by the Max Planck Society.
Footnotes
Publication number 3877 of The Netherlands Institute of Ecology (NIOO-KNAW).
REFERENCES
- 1.Acimovic, Y., and I. R. Coe. 2002. Molecular evolution of the equilibrative nucleoside transporter family: identification of novel family members in prokaryotes and eukaryotes. Mol. Biol. Evol. 19:2199-2210. [DOI] [PubMed] [Google Scholar]
- 2.Alm, E. W., D. B. Oerther, N. Larsen, D. A. Stahl, and L. Raskin. 1996. The oligonucleotide probe database. Appl. Environ. Microbiol. 62:3557-3559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alm, E. W., and D. A. Stahl. 2000. Critical factors influencing the recovery and integrity of rRNA extracted from environmental samples: use of an optimized protocol to measure depth-related biomass distribution in freshwater sediments. J. Microbiol. Methods 40:153-162. [DOI] [PubMed] [Google Scholar]
- 4.Amann, R. I., B. J. Binder, R. J. Olson, S. W. Chisholm, R. Devereux, and D. A. Stahl. 1990. Combination of 16S ribosomal RNA-targeted oligonucleotide probes with flow cytometry for analyzing mixed microbial populations. Appl. Environ. Microbiol. 56:1919-1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baker, B. J., P. Hugenholtz, S. C. Dawson, and J. F. Banfield. 2003. Extremely acidophilic protists from acid mine drainage host Rickettsiales-lineage endosymbionts that have intervening sequences in their 16S rRNA genes. Appl. Environ. Microbiol. 69:5512-5518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bergin, C. 2002. Charakterisierung von stabilen Kohlenstoffisotopen in Gewebe und RNA von Muscheln mit endosymbiontischen Bakterien. Diplomarbeit. Ernst-Moritz-Arndt-Universität, Greifswald, Germany.
- 7.Bergin, C., P. R. Dando, N. Dubilier, and B. J. MacGregor. 2003. Stable carbon isotopic composition of RNA and SSU rRNA in the hydrothermal vent mussel Bathymodiolus azoricus harboring a dual endosymbiont population. Presented at the ASLO Aquatic Sciences Meeting, Salt Lake City, Utah.
- 8.Beste, D. J. V., J. Peters, T. Hooper, C. Avignone-Rossa, M. E. Bushell, and J. McFadden. 2005. Compiling a molecular inventory for Mycobacterium bovis BCG at two growth rates: evidence for growth rate-mediated regulation of ribosome biosynthesis and lipid metabolism. J. Bacteriol. 187:1677-1684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Biagini, G. A., A. J. Rutter, B. J. Finlay, and D. Lloyd. 1998. Lipids and lipid metabolism in the microaerobic free-living diplomonad Hexamita sp. Eur. J. Protistol. 34:148-152. [Google Scholar]
- 10.Boschker, H. T. S. 2004. Linking microbial community structure and functioning: stable isotope (13C) labeling in combination with PLFA analysis, p. 1673-1688. In G. A. Kowalchuk, F. J. de Bruijn, I. M. Head, A. D. Akkermans, and J. D. van Elsas (ed.), Microbial ecology. Kluwer Academic Publishers, Dordrecht, The Netherlands.
- 11.Boschker, H. T. S., W. de Graaf, M. Köster, L.-A. Meyer-Reil, and T. E. Cappenberg. 2001. Bacterial populations and processes involved in acetate and propionate consumption in anoxic brackish sediment. FEMS Microbiol. Ecol. 35:97-103. [DOI] [PubMed] [Google Scholar]
- 12.Boschker, H. T. S., and J. J. Middelburg. 2002. Stable isotopes and biomarkers in microbial ecology. FEMS Microbiol. Ecol. 40:85-95. [DOI] [PubMed] [Google Scholar]
- 13.Boschker, H. T. S., S. C. Nold, P. Wellsbury, D. Bos, W. de Graaf, R. Pel, R. J. Parkes, and T. E. Cappenberg. 1998. Direct linking of microbial populations to specific biogeochemical processes by 13C-labelling of biomarkers. Nature 392:801-805. [Google Scholar]
- 14.Bowman, J. P., and R. D. McCuaig. 2003. Biodiversity, community structural shifts, and biogeography of prokaryotes within Antarctic continental shelf sediment. Appl. Environ. Microbiol. 69:2463-2483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brinch-Iversen, J., and G. M. King. 1990. Effects of substrate concentration, growth state, and oxygen availability on relationships among bacterial carbon, nitrogen and phospholipid phosphorus content. FEMS Microbiol. Ecol. 74:345-355. [Google Scholar]
- 16.Buchan, A., S. Y. Newell, M. Butler, E. J. Biers, J. T. Hollibaugh, and M. A. Moran. 2003. Dynamics of bacterial and fungal communities on decaying salt marsh grass. Appl. Environ. Microbiol. 69:6676-6687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bull, I. D., N. R. Parekh, G. H. Hall, P. Ineson, and R. P. Evershed. 2000. Detection and classification of atmospheric methane oxidizing bacteria in soil. Nature 405:175-178. [DOI] [PubMed] [Google Scholar]
- 18.Canfield, D. E., E. Kristensen, and B. Thamdrup. 2005. Aquatic geomicrobiology, vol. 48. Advances in Marine Biology. Elsevier, Oxford, United Kingdom. [DOI] [PubMed]
- 19.Cox, R. A. 2004. Quantitative relationships for specific growth rates and macromolecular compositions of Mycobacterium tuberculosis, Streptomyces coelicolor A3(2) and Escherichia coli B/r: an integrative theoretical approach. Microbiology 150:1413-1426. [DOI] [PubMed] [Google Scholar]
- 20.Daims, H., A. Bruhl, R. Amann, K. H. Schleifer, and M. Wagner. 1999. The domain-specific probe EUB338 is insufficient for the detection of all bacteria: development and evaluation of a more comprehensive probe set. Syst. Appl. Microbiol. 22:434-444. [DOI] [PubMed] [Google Scholar]
- 21.de Koning, H., and G. Diallinas. 2000. Nucleobase transporters (review). Mol. Membr. Biol. 17:75-94. [DOI] [PubMed] [Google Scholar]
- 22.Dell'Anno, A., M. Fabiano, G. C. A. Duineveld, A. Kok, and R. Danovaro. 1998. Nucleic acid (DNA, RNA) quantification and RNA/DNA ratio determination in marine sediments: comparison of spectrophotometric, fluorometric, and high-performance liquid chromatography methods and estimation of detrital DNA. Appl. Environ. Microbiol. 64:3238-3245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.de Vries, J., and W. Wackernagel. 2004. Microbial horizontal gene transfer and the DNA release from transgenic crop plants. Plant Soil 266:91-104. [Google Scholar]
- 24.Dubnau, D. 1999. DNA uptake in bacteria. Annu. Rev. Microbiol. 53:217-244. [DOI] [PubMed] [Google Scholar]
- 25.Fang, J. S., C. Kato, T. Sato, O. Chan, and D. McKay. 2004. Biosynthesis and dietary uptake of polyunsaturated fatty acids by piezophilic bacteria. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 137:455-461. [DOI] [PubMed] [Google Scholar]
- 26.Fegatella, F., J. Lim, S. Kjelleberg, and R. Cavicchioli. 1998. Implications of rRNA operon copy number and ribosome content in the marine oligotrophic ultramicrobacterium Sphingomonas sp. strain RB2256. Appl. Environ. Microbiol. 64:4433-4438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Flärdh, K., P. S. Cohen, and S. Kjelleberg. 1992. Ribosomes exist in large excess over the apparent demand for protein synthesis during carbon starvation in marine Vibrio sp. strain CCUG 15956. J. Bacteriol. 174:6780-6788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Freitag, T. E., and J. I. Prosser. 2003. Community structure of ammonia-oxidizing bacteria within anoxic marine sediments. Appl. Environ. Microbiol. 69:1359-1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fuchs, B. M., F. O. Glöckner, J. Wülf, and R. Amann. 2000. Unlabeled helper oligonucleotides increase the in situ accessibility to 16S rRNA of fluorescently labeled oligonucleotide probes. Appl. Environ. Microbiol. 66:3603-3607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gallagher, E., L. McGuinness, C. Phelps, L. Y. Young, and L. J. Kerkhof. 2005. 13C-carrier DNA shortens the incubation time needed to detect benzoate-utilizing denitrifying bacteria by stable-isotope probing. Appl. Environ. Microbiol. 71:5192-5196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Godin, J.-P., J. Hau, L.-B. Fay, and G. Hopfgartner. 2005. Isotope ratio monitoring of small molecules and macromolecules by liquid chromatography coupled to isotope ratio mass spectrometry. Rapid Commun. Mass Spectrom. 19:2689-2698. [DOI] [PubMed] [Google Scholar]
- 32.Hanson, J. R., J. L. Macalady, D. Harris, and K. M. Scow. 1999. Linking toluene degradation with specific microbial populations in soil. Appl. Environ. Microbiol. 65:5403-5408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Head, I. M., J. R. Saunders, and R. W. Pickup. 1998. Microbial evolution, diversity, and ecology: a decade of ribosomal RNA analysis of uncultivated microorganisms. Microb. Ecol. 35:1-21. [DOI] [PubMed] [Google Scholar]
- 34.Hicks, R. E., R. I. Amann, and D. A. Stahl. 1992. Dual staining of natural bacterioplankton with 4′,6-diamidino-2-phenylindole and fluorescent oligonucleotide probes targeting kingdom-level 16S ribosomal RNA sequences. Appl. Environ. Microbiol. 58:2158-2163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jørgensen, B. B. 1978. Comparison of methods for the quantification of bacterial sulfate reduction in coastal marine sediments. I. Measurement with radiotracer techniques. Geomicrobiol. J. 1:11-27. [Google Scholar]
- 36.Ju, Q. D., and J. R. Warner. 1994. Ribosome synthesis during the growth cycle of Saccharomyces cerevisiae. Yeast 10:151-157. [DOI] [PubMed] [Google Scholar]
- 37.Kemp, P. F., and J. Y. Aller. 2004. Bacterial diversity in aquatic and other environments: what 16S rDNA libraries can tell us. FEMS Microbiol. Ecol. 47:161-177. [DOI] [PubMed] [Google Scholar]
- 38.Kerkhof, L., and P. Kemp. 1999. Small ribosomal RNA content in marine proteobacteria during non-steady-state growth. FEMS Microbiol. Ecol. 30:253-260. [DOI] [PubMed] [Google Scholar]
- 39.Krummen, M., A. W. Hilkert, D. Juchelka, A. Duhr, H. J. Schlüter, and R. Pesch. 2004. A new concept for isotope ratio monitoring liquid chromatography/mass spectrometry. Rapid Commun. Mass Spectrom. 18:2260-2266. [DOI] [PubMed] [Google Scholar]
- 40.Lim, E. L., L. A. Amaral, D. A. Caron, and E. F. DeLong. 1993. Application of ribosomal RNA-based probes for observing marine nanoplanktonic protists. Appl. Environ. Microbiol. 59:1647-1655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ludwig, W., O. Strunk, R. Westram, L. Richter, H. Meier, Yadhukumar, A. Buchner, T. Lai, S. Steppi, G. Jobb, W. Förster, I. Brettske, S. Gerber, A. W. Ginhart, O. Gross, S. Grumann, S. Hermann, R. Jost, A. König, T. Liss, R. Lüßmann, M. May, B. Nonhoff, B. Reichel, R. Strehlow, A. Stamatakis, N. Stuckmann, A. Vilbig, M. Lenke, T. Ludwig, A. Bode, and K.-H. Schleifer. 2004. ARB: a software environment for sequence data. Nucleic Acids Res. 32:1363-1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lueders, T., M. Manefield, and M. W. Friedrich. 2004. Enhanced sensitivity of DNA- and rRNA-based stable isotope probing by fractionation and quantitative analysis of isopycnic centrifugation gradients. Environ. Microbiol. 6:73-78. [DOI] [PubMed] [Google Scholar]
- 43.Lyons, J. I., S. Y. Newell, R. P. Brown, and M. A. Moran. 2005. Screening for bacterial-fungal associations in a south-eastern US salt marsh using pre-established fungal monocultures. FEMS Microbiol. Ecol. 54:179-187. [DOI] [PubMed] [Google Scholar]
- 44.MacGregor, B. J., V. Brüchert, S. Fleischer, and R. Amann. 2002. Isolation of small-subunit rRNA for stable isotopic characterization. Environ. Microbiol. 4:451-464. [DOI] [PubMed] [Google Scholar]
- 45.MacGregor, B. J., and D. Juchelka. 2005. Determining stable carbon isotopic composition of rRNA by HPLC-IRMS: a new method to discover who is eating what in microbial communities. Presented at the ASLO Aquatic Sciences Meeting, Salt Lake City, Utah.
- 46.MacGregor, B. J., D. P. Moser, E. W. Alm, K. H. Nealson, and D. A. Stahl. 1997. Crenarchaeota in Lake Michigan sediment. Appl. Environ. Microbiol. 63:1178-1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Manefield, M., A. S. Whiteley, R. I. Griffiths, and M. J. Bailey. 2002. RNA stable isotope probing, a novel means of linking microbial community function to phylogeny. Appl. Environ. Microbiol. 68:5367-5373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Middelburg, J. J., C. Barranguet, H. T. S. Boschker, P. M. J. Herman, T. Moens, and C. H. R. Heip. 2000. The fate of intertidal microphytobenthos carbon: an in situ 13C-labeling study. Limnol. Oceanog. 45:1224-1234. [Google Scholar]
- 49.Mulder, M. M., H. M. L. van der Gulden, P. W. Postma, and K. van Dam. 1988. Effect of macromolecular composition of microorganisms on the thermodynamic description of their growth. Biochim. Biophys. Acta 936:406-412. [DOI] [PubMed] [Google Scholar]
- 50.Neidhardt, F. C., and H. E. Umbarger. 2004, posting date. Chemical composition of E. coli. In Escherichia coli and Salmonella: cellular and molecular biology. American Society for Microbiology, Washington, D.C. [Online.] http://www.ecosal.org/ecosal/index.jsp.
- 51.Nelson, D. L., and M. M. Cox. 2004. Lehninger principles of biochemistry. W. H. Freeman, New York, N.Y.
- 52.Pearson, A., A. L. Sessions, K. J. Edwards, and J. M. Hayes. 2004. Phylogenetically specific separation of rRNA from prokaryotes for isotopic analysis. Mar. Chem. 92:295-306. [Google Scholar]
- 53.Pelz, O., M. Tesar, R. M. Wittich, E. R. B. Moore, K. N. Timmis, and W. R. Abraham. 1999. Towards elucidation of microbial community metabolic pathways: unraveling the network of carbon sharing in a pollutant-degrading bacterial consortium by immunocapture and isotopic ratio mass spectrometry. Environ. Microbiol. 1:167-174. [DOI] [PubMed] [Google Scholar]
- 54.Pernthaler, A., J. Pernthaler, H. Eilers, and R. Amann. 2001. Growth patterns of two marine isolates: adaptations to substrate patchiness? Appl. Environ. Microbiol. 67:4077-4083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Radajewski, S., P. Ineson, N. R. Parekh, and J. C. Murrell. 2000. Stable-isotope probing as a tool in microbial ecology. Nature 403:646-649. [DOI] [PubMed] [Google Scholar]
- 56.Ravenschlag, K., K. Sahm, and R. Amann. 2001. Quantitative molecular analysis of the microbial community in marine Arctic sediments (Svalbard). Appl. Environ. Microbiol. 67:387-395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ravenschlag, K., K. Sahm, J. Pernthaler, and R. Amann. 1999. High bacterial diversity in permanently cold marine sediments. Appl. Environ. Microbiol. 65:3982-3989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rosset, R., J. Julien, and R. Monier. 1966. Ribonucleic acid composition of bacteria as a function of growth rate. J. Mol. Biol. 18:308-320. [DOI] [PubMed] [Google Scholar]
- 59.Schaechter, M., O. Maaløe, and N. O. Kjeldgaard. 1958. Dependency on medium and temperature of cell size and chemical composition during balanced growth of Salmonella typhimurium. J. Gen. Microbiol. 19:592-606. [DOI] [PubMed] [Google Scholar]
- 60.Sessions, A. L., S. P. Sylva, and J. M. Hayes. 2005. Moving-wire device for carbon isotopic analyses of nanogram quantities of nonvolatile organic carbon. Anal. Chem. 77:6519-6527. [DOI] [PubMed] [Google Scholar]
- 61.Stahl, D. A., B. Flesher, H. R. Mansfield, and L. Montgomery. 1988. Use of phylogenetically based hybridization probes for studies of ruminal microbial ecology. Appl. Environ. Microbiol. 54:1079-1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Stoeck, T., and S. Epstein. 2003. Novel eukaryotic lineages inferred from small-subunit rRNA analyses of oxygen-depleted marine environments. Appl. Environ. Microbiol. 69:2657-2663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tunlid, A., and D. C. White. 1992. Biochemical analysis of biomass, community structure, nutritional status, and metabolic activity of microbial communities in soil, p. 229-262. In J. M. Bollag and G. Stotzky (ed.), Soil biochemistry. Marcel Dekker, New York, N.Y.
- 64.Vrede, T., D. R. Dobberfuhl, S. A. L. M. Kooijman, and J. J. Elser. 2004. Fundamental connections among organism C:N:P stoichiometry, macromolecular composition, and growth. Ecology 85:1217-1229. [Google Scholar]
- 65.Watanabe, K., C. Ishikawa, H. Inoue, D. Cenhua, K. Yazawa, and K. Kondo. 1994. Incorporation of exogenous docosahexaenoic acid into various bacterial lipids. J. Am. Oil Chem. Soc. 71:325-330. [Google Scholar]