Abstract
Microbiologists conducting surveys of bacterial and archaeal diversity often require comparative alignments of thousands of 16S rRNA genes collected from a sample. The computational resources and bioinformatics expertise required to construct such an alignment has inhibited high-throughput analysis. It was hypothesized that an online tool could be developed to efficiently align thousands of 16S rRNA genes via the NAST (Nearest Alignment Space Termination) algorithm for creating multiple sequence alignments (MSA). The tool was implemented with a web-interface at http://greengenes.lbl.gov/NAST. Each user-submitted sequence is compared with Greengenes' ‘Core Set’, comprising ∼10 000 aligned non-chimeric sequences representative of the currently recognized diversity among bacteria and archaea. User sequences are oriented and paired with their closest match in the Core Set to serve as a template for inserting gap characters. Non-16S data (sequence from vector or surrounding genomic regions) are conveniently removed in the returned alignment. From the resulting MSA, distance matrices can be calculated for diversity estimates and organisms can be classified by taxonomy. The ability to align and categorize large sequence sets using a simple interface has enabled researchers with various experience levels to obtain bacterial and archaeal community profiles.
INTRODUCTION
DNA sequence information from the 1.5 kb small subunit 16S ribosomal RNA (rRNA) gene has been used to successfully identify and phylogenetically classify microorganisms from environmental and medical samples (1–3). In more ambitious efforts, the relative abundance of bacterial groups has been estimated by sequencing hundreds to thousands of 16S rRNA genes derived from a sample (4–9). However, a bottleneck in data analysis is encountered in creating multiple sequence alignments (MSA). The MSA is a common means of communicating a proposed positional homology among many genes using a column-by-column format. It can be stored and presented in a variety of formats but in all cases it represents a two-dimensional matrix with each row describing a gene and each column holding the nucleotide found at a certain position along the gene. Alignments are useful when gaps have been appropriately added to mark an inference of an insertion or deletion event where one sequence has a base while another sequence lacks a base at the corresponding position. This process yields sequence strings occupying an equal number of columns allowing the matrix to form a true rectangle. MSAs are desirable for annotation of conserved versus variable gene loci by observing heterogeneity along the columns, recruiting columns with sufficient data for inter-row (sequence) comparison, and calculating distance matrices for row clustering. ClustalW (10) is a commonly used progressive MSA method for inserting gaps into sequences to achieve perfect rectangles. Hundreds of diverse sequences can be aligned using this approach to establish a ‘profile’ alignment. Later, new sequences can be added to this profile without re-computing the optimal gap placements for the entire alignment.
Frequently, when adding a candidate sequence to a MSA profile, one or more internal insertions will be discovered which cannot be accommodated in the profile. This event requires a researcher to make one of two choices: (i) allow the column count to grow whenever an insertion is required, which requires each sequence to gain more characters or (ii) allow a local misalignment within a sequence (row) so that the insertion does not disrupt the entire alignment format of the profile. Until now, the choice readily available has been the former as implemented by ClustalW. Certain objectives are left unsatisfied by this approach. In some instances, the apparent need to create new columns in the MSA owes to the presence of poor quality sequences. If allowed, the MSA could expand to a cumbersome collection of unsubstantiated insertion inferences (gaps). Ongoing comparative sequence analysis projects benefit from having a fixed column count in the MSA, enabling unchanging annotation of position-dependent features such as primer annealing locations, secondary structures and column masks. Furthermore, collaborative MSA construction becomes problematic when copies of a single profile diverge in column content as individual researchers add their own unique data. To enable fixed column counts, allow piecemeal MSA curation and support collaborations in comparative genomics the local misalignment approach is now available and implemented via NAST (Nearest Alignment Space Termination).
We have established a web service for creating NAST MSAs from user data which is intended to facilitate comparison of 16S rRNA gene sequences from bacteria and archaea. This service has performed well in aligning thousands of user-supplied sequences into a single MSA while optionally intercalating genes from reference organisms. It was created to handle large datasets produced in exploratory microbial ecology, medical microbiology and metagenomics. One unique feature is that NAST can output the MSA in a standard, consistent format of 7682 characters per sequence so that similar loci are located at dependable positions from batch to batch (necessary for large, ongoing projects). An optional pre-processing of data based on chromatogram quality scores is allowed and post-processing options include distance matrix creation and taxonomic classification using five independent curators' nomenclature.
We have received considerable positive feedback from diverse users who have collectively submitted over 1600 jobs.
NAST ALGORITHM
The first version of the alignment compression algorithm, NAST (11), was designed to produce uniform MSAs of 16S rRNA genes obtained from public repositories. The current version contains improvements and has been made available via a web-interface for alignment of pre-published collections.
A current set of >80 000 16S rDNA genes gleaned from GenBank is maintained in aligned format on the Greengenes server (12). From this collection, a smaller, high-quality reference group was sought. The size of this group is a consequence of balancing both the need to encompass the full set's diversity and the application's requirement of rapid searching. Similarity comparison for one sequence against 105 can occur in a reasonable time period for an online tool. After clustering the full set with a sliding scale of similarity, it was found that a 96% identity threshold produced a cluster count of the ideal magnitude. From each cluster, one sequence record was chosen by favoring long gene sequences with low nucleotide ambiguity from published microbial isolates according to the default weighting in the de-replication tool (http://greengenes.lbl.gov/Derep). The resulting ‘Core Set’ of 10 270 aligned records were non-chimeric (13) and >1250 ntin length. The terminal ends of incomplete sequences (with lengths between 1250 and 1500 nt) were imputed from known sequence data of near-neighbors. The projected termini of the sequences in the Core Set are only used as a reference for the NAST alignment tool and are not entered into the Greengenes database. The Core Set is considered the profile MSA and consists of the template sequences aligned into 7682 columns.
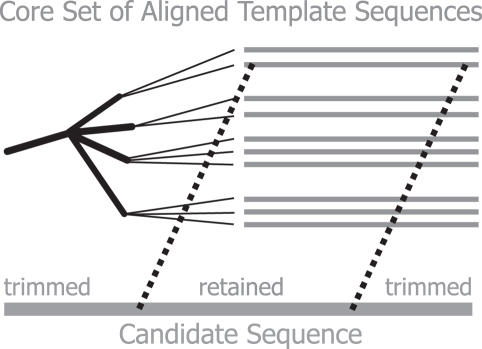
An unaligned sequence is termed the ‘candidate’ and is matched to templates by comparison of 7mers in common (Figure 1). A BLAST (14) alignment with parameter ‘q = −1' is performed to pair bases from candidate to template. BioPerl (15) parsers are used for extracting detail from the BLAST report. To eliminate extra-16S rRNA sequence, the candidate sequence is trimmed to that which is bound by the beginning and end points of a single BLAST alignment span. Although this process will fractionate chimeras where the candidate is composed of sequence fragments from vastly dissimilar organisms, NAST should not be regarded as a substitution for dedicated chimera detection software. Finally, the trimmed candidate is reverse complemented whenever opposite strands from the subject and query are paired.
Figure 1.
Locating a NAST alignment template for a user-supplied candidate sequence. Candidate sequence in green is matched to a near-neighbor aligned template in Greengenes' Core Set (grey) by tallying 7mers in common. The alignment ‘template’ is BLAST aligned to the candidate parameter q = −1 (favors long match). The candidate is then trimmed of flanking sequence data such as tRNA, intergenic spacer regions, vector sequence, 23S rDNA and sequence outside of the high-scoring pair (HSP) boundaries. If the HSP pairs opposite strands, then the candidate is reverse complemented.
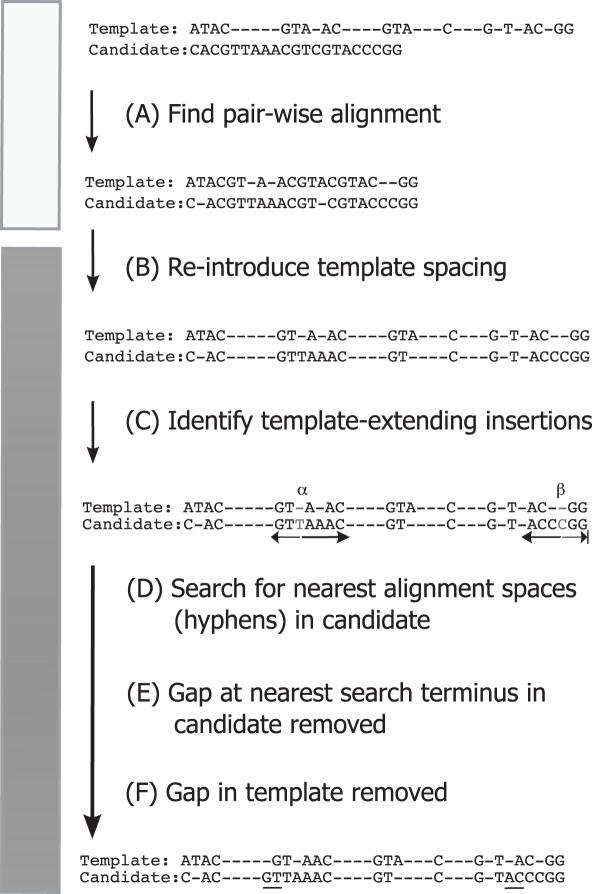
As a result of the pairwise alignment performed by BLAST, new alignment gaps (hyphens) are introduced between the bases of the template whenever the candidate contains additional internal bases (insertions) compared with the template (Figure 2A, B). Any pairwise alignment algorithm must do this to compensate for nucleotides not shared by both sequences. This expansion, when intercalated with the original template spacing, results in candidates occupying more columns (characters) than the original template format (Figure 2C). Since a consistent column count may be an option chosen by the user, the candidate-template alignment is compressed back to 7682 characters with NAST. After insertion bases are identified (Figure 2C), a bi-directional search for the nearest alignment space (hyphen) relative to the insertion results in character deletion of the proximal place holders. Ultimately, local misalignments, spanning from the insertion base to the deleted alignment space, are permitted to preserve the global MSA format.
Figure 2.
Example of NAST compression of a BLAST pairwise alignment using a 38 character aligned template. Template and candidate is extended to 40 characters after (A) BLAST gap insertion and (B) retention of original template spacing. (C) Nucleotide insertions in the candidate relative to the template which force additional characters to be added in the template are identified at positions α and β. (D) A bi-directional search for the nearest alignment space (hyphen) relative to the insertion terminates at the positions indicated by the black arrows. The leftward search from the α position was shorter in distance compared with the rightward, thus the space to the left of ‘GT’ was removed. (E) The search from the β position encountered the alignment edge on the right, thus the position to the left of ‘AC’ was removed. (F) Lastly, the two template-extending spaces are deleted from the template. Notice that sequence data are not added to or overwritten in the candidate. The NAST removal of two characters from both sequences allowed local misalignments (underlined) while preserving the 38 character format of the global MSA.
NAST WEB SERVER
Submitting Jobs
Instructions for using the NAST aligner as well as other Greengenes tools are available in the tutorial at http://greengenes.lbl.gov/Download/Tutorial/. In brief, batches of sequences are submitted in FASTA format. Users can select the minimum candidate-template similarity in terms of length and percent identity to constrain the NAST alignment from attempting to align records that may not be 16S rRNA gene sequences. Users may also opt to have near-neighbor sequences from the Core Set added to the resulting MSA. Formatting options allow either all columns to be returned (each returned sequence will be exactly 7682 characters long) or removal of common alignment gap characters (returned sequences will contain an equal number of characters ≤7682, all columns containing only place holders will be removed). Lastly, users choose the output format from a list that includes FASTA, ClustalW, MEGA (16), PHYLIP (17) and others.
Result Files
Two files are returned to the user by Email. The MSA is delivered as a compressed document and a summary table is sent as a tab-delimitated text file (Table 1). The summary describes the fate of each sequence in the submitted batch. If a sequence diverges from the Core Set beyond the user's thresholds, then an informative error message is reported. Otherwise, the following information is returned: candidate's sequence length as submitted, a Greengenes sequence identifier for the template, the longest nucleotide insertion relative to template and the post-NAST nucleotide count. Comparing the submitted length with the post-NAST length can alert the user of unexpected sequence truncation. Minor truncation of one to five bases occurs when terminal bases cannot be accurately aligned. Large truncations indicate that either non-gene data were in the record or that BLAST found matches distributed to multiple Core Set sequences, possibly suggesting chimeric content. Long insertions are reported for identification of sequences divergent from the Core Set.
Table 1.
Example summary of NAST output describing the fate of each sequence in the submitted batch. This report is delivered with the MSA
| Candidate sequence ID | Candidate nucleotide count | Errors | Template IDa | BLAST percent identity to template | Longest insertion relative to template | Candidate span aligned | Candidate nucleotide count post-NAST |
|---|---|---|---|---|---|---|---|
| AKIW1006 | 1475 | 137800 | 96.81 | 6 | 1..1475 | 1475 | |
| AKIW1010 | 1486 | 85092 | 94.90 | 0 | 1..1486 | 1486 | |
| AKIW1012 | 1458 | 44738 | 86.50 | 8 | 1..1458 | 1458 | |
| AKIW1013 | 1423 | 137999 | 100.00 | 0 | 1..1423 | 1423 | |
| AKIW1030 | 1513 | 14808 | 98.94 | 0 | 3..1513 | 1511 | |
| AKIW448 | 1462 | Xb | |||||
| AKIW474 | 1473 | 134781 | 99.12 | 1 | 1..1473 | 1473 | |
| AE017333.1 | 4222645 | 75819 | 99.42 | 1 | 920782..922330 | 1549 | |
| AE017333.1 | 4222645 | 75819 | 99.42 | 1 | 158106..159654 | 1549 | |
| AE017333.1 | 4222645 | 75819 | 99.48 | 0 | 611802..613349 | 1548 | |
| AE017333.1 | 4222645 | 75819 | 99.42 | 0 | 9710..11257 | 1548 | |
| AE017333.1 | 4222645 | 75819 | 99.35 | 1 | 95150..96698 | 1549 | |
| AE017333.1 | 4222645 | 75819 | 99.35 | 1 | complement (3121607..3123155) | 1549 | |
| AE017333.1 | 4222645 | 75819 | 99.55 | 0 | 34408..35955 | 1548 |
aTemplate ID is an unique numerical identifier for a Greengenes 16S rRNA gene record.
bError column warns when the individual sequence alignment quality did not meet the user-defined thresholds. This table cell contains the text ‘Length:1238 Percent Indentity:93.21 Template:137857’. Error generated due to user’s requirement that each candidate sequence aligned to a template sequence along at least 1250 bases.
Performance
With the current Greengenes hardware configuration using Intel Xeon 2.4GHz processors, NAST is able to align ∼10 16S rRNA gene sequences per minute. Since each sequence in a batch does not require comparisons with all other batched sequences, the method scales linearly. Pre-release users from five research institutions verified that the NAST web server reliably returned aligned sequences in a timely manner, but cautioned that returned alignment files can be large and may not be accepted by some Email servers. In all cases, breaking the batch into sub-sets overcame the constraint.
Suggested strategy for microbial community assessment using NAST
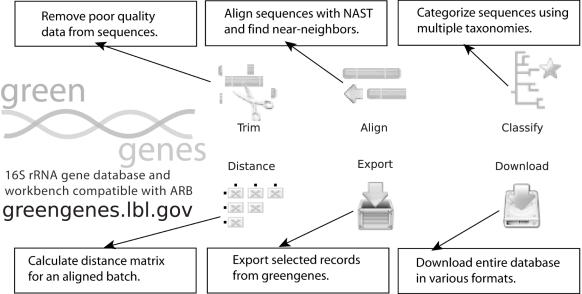
Greengenes supplies not only an aligned 16S rRNA gene reference database but also maintains a suite of sequence analysis tools (Figure 3). In the typical scenario, a researcher obtains a complex pool of 16S rRNA genes from a variety of bacterial genomes present in an environmental or medical sample. The DNA is serially sampled by cloning and sequencing. The raw sequencing reads can be trimmed of low quality terminal fragments using the ‘Trim’ tool following phred (18) chromatogram scoring. The NAST tool is then used to create the MSA and maintain the 7682-character format. Once aligned, the entire batch can be classified by Greengenes using taxonomic nomenclature proposed by independent curators [NCBI, RDP/Bergey's (19) Ludwig (20) Hugenholtz (21) and Pace (22)]. Since more than one estimation of phylogenetic descent is returned, the user can implement a balanced approach for node nomenclature when generating project-specific dendrograms. In addition, Greengenes is able to calculate a distance matrix using PHYLIP's DNADIST providing a suitable input format for construction of collector's and rarefaction curves using DOTUR (23) while also permitting de novo tree plotting. Combining pertinent public records into a project can be accomplished automatically at the NAST step or advanced users can export individual records or the entire Greengenes database in multiple popular formats including ARB (20), MEGA and FASTA.
Figure 3.
Greengenes pre-processing and post-processing tools for use with the NAST aligner. ‘Trim’ can be used to remove poor quality DNA data before alignment. ‘Classify’ and ‘Distance’ receive NAST MSAs as input. ‘Export’ and ‘Download’ allow advanced users to append their MSA with select sequences from the public repositories.
APPLICATIONS
The NAST web service has already been used to process data for a recent publication and several manuscripts currently in preparation or review. Exploring the diversity of bacterial symbionts found in vertebrate digestive tracts has required massive MSAs. The NAST web server was used to align and compare 16S rRNA genes recovered from the gut of fish, humans and mice to those found in the open ocean (24). An extended investigation covering the intestinal microbiota of 30 mammalian species aligned ∼20 000 16S rRNA gene sequences (R. E. Ley, P. Turnbaugh and J. I. Gordon, unpublished data). The NAST approach was also utilized to align and subsequently categorize 4023 feces-derived 16S rDNA sequences obtained as part of a longitudinal study of the human neonatal GI tract (C. Palmer, P. O. Brown, E. M. Bik and D. A. Relman, unpublished data).
Environmental microbial sampling has produced sizable sets of 16S rRNA gene sequences as well. An 1800 sequence set from uranium contaminated soil, deep sub-surface water, and urban aerosols was NAST aligned, allowing analysis of sample diversity as well as evaluation of parallel 16S rRNA microarray results (T. Z. DeSantis, E. L. Brodie, Y. M. Piceno and G. L. Andersen, manuscript in review). To uncover novel 16S rRNA types in an environmental sample, short ‘miniprimers’ (∼10 nt) were tested to expand the scope of recoverable by PCR sequences and although the amplicons were unique from existing Greengenes database entries, NAST alignment was successful, facilitating import into ARB for phylogenetic categorization (T. A. Isenbarger, M. Finney, J. Handelsman and G. Ruvkun, in preparation). A comprehensive annotation of 18 000 GenBank 16S rRNA genes from natural environments has also been made possible with NAST (C. Lozupone and R. Knight, unpublished data).
FUTURE DEVELOPMENT
Since the NAST approach relies on a well-aligned profile of diverse sequences we endeavor to make manual improvements to the Core Set as needed. We are aware that research groups who rely upon accurate 16S rRNA gene comparisons are regularly curating individual sequence alignments. Since these efforts can benefit other users, we conceive a conduit for transmitting the improvements to interested parties. When manifest, users will be able to not only suggest gap-placement alterations in the Core Set or other publicly distributed sequences but also to recommend sequence additions to the Core Set as new levels of microbial diversity are unearthed.
To extend the applicability of the NAST aligner for use with other standard 16S rRNA gene alignment formats we plan to add options for building MSAs with Ludwig or RDP column positioning. Also, a stand-alone version of the NAST software is in development. This will allow installation of NAST on a laboratory computer/server for more rapid analysis and customization and will enable the local curation of MSAs in Greengenes' or other formats. In theory, NAST's utility is not limited to 16S rRNA data. Sizeable MSAs of other genes or proteins, such as those encoding 18S rRNA, rpoB or recA can be built and maintained. Sequences can be merged into any existing MSA profile providing the trade-off between fixed total alignment string length and the extent of local misalignment is acceptable.
CONCLUSIONS
The NAST web server is available for creating MSAs of small and large 16S rRNA gene sequencing projects. NAST allows retention of fixed MSA column counts regardless of the quantity of records added to a profile alignment. This permits ongoing MSA curation, and supports collaborative efforts in comparative genomics. NAST and supporting tools at the Greengenes website enable microbiologists to rapidly compare sampled sequences to publicly available reference sequences as well as to each other.
Acknowledgments
The computational infrastructure was provided in part by the Virtual Institute for Microbial Stress and Survival (http://VIMSS.lbl.gov) supported by the US Department of Energy, Office of Science, Office of Biological and Environmental Research, Genomics:GTL Program and the Natural and Accelerated Bioremediation Research Program through contract DE-AC02-05CH11231 between Lawrence Berkeley National Laboratory and the US Department of Energy. Web application development was funded in part by the Department of Homeland Security under grant number HSSCHQ04X00037. Funding to pay the Open Access publication charges for this article was provided by the U.S. Department of Homeland Security.
Conflict of interest statement. None declared.
REFERENCES
- 1.Fox G.E., Stackebrandt E., Hespell R.B., Gibson J., Maniloff J., Dyer T.A., Wolfe R.S., Balch W.E., Tanner R.S., Magrum L.J., et al. The phylogeny of prokaryotes. Science. 1980;209:457–463. doi: 10.1126/science.6771870. [DOI] [PubMed] [Google Scholar]
- 2.Woese C.R., Fox G.E., Zablen L., Uchida T., Bonen L., Pechman K., Lewis B.J., Stahl D. Conservation of primary structure in 16S ribosomal RNA. Nature. 1975;254:83–86. doi: 10.1038/254083a0. [DOI] [PubMed] [Google Scholar]
- 3.Kong Y., Ong S.L., Ng W.J., Liu W.T. Diversity and distribution of a deeply branched novel proteobacterial group found in anaerobic–aerobic activated sludge processes. Environ. Microbiol. 2002;4:753–757. doi: 10.1046/j.1462-2920.2002.00357.x. [DOI] [PubMed] [Google Scholar]
- 4.Hughes J.B., Hellmann J.J., Ricketts T.H., Bohannan B.J. Counting the uncountable: statistical approaches to estimating microbial diversity. Appl. Environ. Microbiol. 2001;67:4399–4406. doi: 10.1128/AEM.67.10.4399-4406.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Eckburg P.B., Bik E.M., Bernstein C.N., Purdom E., Dethlefsen L., Sargent M., Gill S.R., Nelson K.E., Relman D.A. Diversity of the human intestinal microbial flora. Science. 2005;308:1635–1638. doi: 10.1126/science.1110591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ley R.E., Backhed F., Turnbaugh P., Lozupone C.A., Knight R.D., Gordon J.I. Obesity alters gut microbial ecology. Proc. Natl Acad. Sci. USA. 2005;102:11070–11075. doi: 10.1073/pnas.0504978102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Radosevich J.L., Wilson W.J., Shinn J.H., DeSantis T.Z., Andersen G.L. Development of a high-volume aerosol collection system for the identification of air-borne micro-organisms. Lett. Appl. Microbiol. 2002;34:162–167. doi: 10.1046/j.1472-765x.2002.01048.x. [DOI] [PubMed] [Google Scholar]
- 8.Tringe S.G., von Mering C., Kobayashi A., Salamov A.A., Chen K., Chang H.W., Podar M., Short J.M., Mathur E.J., Detter J.C., et al. Comparative metagenomics of microbial communities. Science. 2005;308:554–557. doi: 10.1126/science.1107851. [DOI] [PubMed] [Google Scholar]
- 9.Griffiths R.I., Whiteley A.S., O'Donnell A.G., Bailey M.J. Physiological and community responses of established grassland bacterial populations to water stress. Appl. Environ. Microbiol. 2003;69:6961–6968. doi: 10.1128/AEM.69.12.6961-6968.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Thompson J.D., Higgins D.G., Gibson T.J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.DeSantis T.Z., Dubosarskiy I., Murray S.R., Andersen G.L. Comprehensive aligned sequence construction for automated design of effective probes (CASCADE-P) using 16S rDNA. Bioinformatics. 2003;19:1461–1468. doi: 10.1093/bioinformatics/btg200. [DOI] [PubMed] [Google Scholar]
- 12.DeSantis T.Z., Hugenholtz P., Larsen N., Rojas M., Brodie E.L., Keller K., Huber T., Dalevi D., Hu P., Andersen G.L. Greengenes: Chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl. Environ. Microbiol. 2006 doi: 10.1128/AEM.03006-05. accepted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huber T., Faulkner G., Hugenholtz P. Bellerophon: a program to detect chimeric sequences in multiple sequence alignments. Bioinformatics. 2004;20:2317–2319. doi: 10.1093/bioinformatics/bth226. [DOI] [PubMed] [Google Scholar]
- 14.Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 15.Stajich J.E., Block D., Boulez K., Brenner S.E., Chervitz S.A., Dagdigian C., Fuellen G., Gilbert J.G., Korf I., Lapp H., et al. The Bioperl toolkit: Perl modules for the life sciences. Genome Res. 2002;12:1611–1618. doi: 10.1101/gr.361602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kumar S., Tamura K., Nei M. MEGA3: Integrated software for Molecular Evolutionary Genetics Analysis and sequence alignment. Brief Bioinform. 2004;5:150–163. doi: 10.1093/bib/5.2.150. [DOI] [PubMed] [Google Scholar]
- 17.Felsenstein J. PHYLIP—phylogeny inference package (Version 3.65) Cladistics. 1989;5:164–166. [Google Scholar]
- 18.Ewing B., Green P. Base-calling of automated sequencer traces using phred. II. Error probabilities. Genome Res. 1998;8:186–194. [PubMed] [Google Scholar]
- 19.Cole J.R., Chai B., Farris R.J., Wang Q., Kulam S.A., McGarrell D.M., Garrity G.M., Tiedje J.M. The Ribosomal Database Project (RDP-II): sequences and tools for high-throughput rRNA analysis. Nucleic Acids Res. 2005;33:D294–D296. doi: 10.1093/nar/gki038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ludwig W., Strunk O., Westram R., Richter L., Meier H., Yadhukumar, Buchner A., Lai T., Steppi S., Jobb G., et al. ARB: a software environment for sequence data. Nucleic Acids Res. 2004;32:1363–1371. doi: 10.1093/nar/gkh293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hugenholtz P. Exploring prokaryotic diversity in the genomic era. Genome Biol. 2002;3:1–8. doi: 10.1186/gb-2002-3-2-reviews0003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pace N.R. A molecular view of microbial diversity and the biosphere. Science. 1997;276:734–740. doi: 10.1126/science.276.5313.734. [DOI] [PubMed] [Google Scholar]
- 23.Schloss P.D., Handelsman J. Introducing DOTUR, a computer program for defining operational taxonomic units and estimating species richness. Appl. Environ. Microbiol. 2005;71:1501–1506. doi: 10.1128/AEM.71.3.1501-1506.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ley R.E., Peterson D.A., Gordon J.I. An extended view of ourselves: ecological and evolutionary forces that shape microbial diversity and genome content in the human intestine. Cell. 2006;124:837–848. doi: 10.1016/j.cell.2006.02.017. [DOI] [PubMed] [Google Scholar]