Abstract
We provide a ‘REMUS’ (reinforced merging techniques for unique peptide segments) web server for identification of the locations and compositions of unique peptide segments from a set of protein family sequences. Different levels of uniqueness are determined according to substitutional relationship in the amino acids, frequency of appearance and biological properties such as priority for serving as candidates for epitopes where antibodies recognize. REMUS also provides interactive visualization of 3D structures for allocation and comparison of the identified unique peptide segments. Accuracy of the algorithm was found to be 70% in terms of mapping a unique peptide segment as an epitope. The REMUS web server is available at http://biotools.cs.ntou.edu.tw/REMUS and the PC version software can be freely downloaded either at http://bioinfo.life.nthu.edu.tw/REMUS or http://spider.cs.ntou.edu.tw/BioTools/REMUS. User guide and working examples for PC version are available at http://spider.cs.ntou.edu.tw/BioTools/REMUS-DOCS.html, and details of the proposed algorithm can be referred to the documents as described previously [H. T. Chang, T. W. Pai, T. C. Fan, B. H. Su, P. C. Wu, C. Y. Tang, C. T. Chang, S. H. Liu and M. D. T. Chang (2006) BMC Bioinformatics, 7, 38 and T. W. Pai, B. H. Su, P. C. Wu, M. D. T. Chang, H. T. Chang, T. C. Fan and S. H. Liu (2006) J. Bioinform. Comput. Biol., 4, 75–92].
INTRODUCTION
A protein family, and its related domains, is defined as a set of proteins that possess a common evolutionary origin reflected by their relationship in function that can usually be observed by sequence homology, or in higher order of structures (1,2). However, some experiments have revealed that enzymes with high sequence identity may possess differential biological functions other than the common catalytic abilities, probably due to the interactions among the variable regions in the sequence and different cellular compartments (3). Therefore, it is informative to identify the localization and compositions of the unique peptide segments in each member of a protein family to correlate with their unique functions.
Although most of the time allocation of major variations among a few query sequences can be achieved by direct multiple sequence alignment, some of the unique peptide segments that are not well aligned will be neglected employing conventional alignment tools. As the amount of various divisions or family protein sequences increases, the task of cross-checking for unique peptide segments becomes difficult. It is quite time consuming and expensive to experimentally search for such unique peptide segments that may involve key biological functions of interest. Thus, an efficient methodology for identifying all unique peptide segments located in a number of query protein sequences is urgently needed. The predicted unique peptide segments serving as sequential epitopes provide applications in designing experiments for specifically differentiating a member from a protein family or characterizing the antibody-recognition sites of protein antigens, and the proposed method can be applied for engineering peptide antigens for the use in immunobiology.
System design and implementation
We present a tool, REMUS, which identifies sequential unique peptide segments within the related protein sequences. Both web interface and PC versions are designed for different user environments, and which can be executed either automatically or manually with respect to the parameter settings (4). REMUS provides efficient identification of unique peptide segments from imported sequences within the widely used FASTA or PDB formats. In addition, REMUS features the substitutable and mismatching characteristics employing flexible approximate matching techniques that provide prediction results in various uniqueness levels for further biological evaluations.
The web version of REMUS system is implemented on IIS with Windows server as an operating system, and the web interface is written in PHP program. Open GL (http://www.opengl.org/) and Jmol (http://jmol.sourceforge.net/) are incorporated as the visualization tools for PC and web version, respectively.
Algorithm
The proposed method employs a sequence-based searching algorithm consisting of a three-phase operation including clustering, searching and merging phases. In the clustering phase, the module classifies 20 amino acids into different groups based on specified BLOSUM/PAM series of matrices (5,6) or simply customized according to user's preference. The traditional hierarchical clustering algorithms and a novel bitwise clustering methodology are both incorporated to enhance the importance of the substitution properties. For example, in the BLOSUM62 substitution matrix, amino acids F and Y possess the substitution scores of 3. If a specified thresholding value of 2 is assigned for clustering processes, then F and Y will be grouped into the same set after the clustering. Therefore, the quantity of predicted unique peptide will be reduced by substitutable properties.
The searching phase performs exact or approximate string matching to extract fundamental unique peptide segments, named as primary pattern. The length of a primary pattern is an important parameter for appropriate unique peptide extraction and strongly influences the final results. The rule of thumb for primary pattern lengths is that a longer length setting for similar sequences and a shorter length setting for dissimilar sequences. To assist users to define the length of primary pattern, an analytical tool based on the percentages of searched unique peptides and non-merged subsegments is provided in the REMUS system. Users can simply select the primary pattern length analysis to obtain a recommended parameter setting prior to searching processes. The searching module performs Boyer Moore algorithm to efficiently retrieve all primary patterns based on previous clustering definitions. Each searched fundamental unique peptide segment will be analyzed based on its frequencies of appearance and its representation level of uniqueness is calculated for the subsequent merging processes.
In the last phase, merging algorithms initiate a novel methodology to extract unique peptide segments by bottom-up merging processes. Four different merging methods, Merge, Trim-Merge, Strict-Merge and Strict-Trim-Merge, are designed to combine the primary unique peptide segments and result in proper subset relationships that reflect the precision of their unique features. The construction of a merged segment w from overlapped primary patterns u and v is formulated in the following equation.
| 1 |
where w(i) is the i-th residue of w, m is the length of u and v, and l is the number of overlapped residues. The criterion for the ‘Merge’ operation is 1 ≤ l < m − 1 and l = m − 1 represents the condition for the ‘Strict-Merge’ case. For the trimming operations, the merged segments are re-examined at the two ends with length of m − 1 and the imperfect residues are eliminated if they appeared in other merged segments. In addition, as the merged unique peptide segments from the query sequences are allocated, the system will rank the identified segments according to four key features characteristic to the antigenic properties of amino acids: hydrophilicity, charge, number of prolines and the proximity of the segments towards the N- or C-terminal end of the protein. A higher ranking indicates better antigenicity in the unique peptide segment. Consequently, segments with a higher ranking may be suggested to serve as a suitable epitope or peptide antigen for generation of a specific antibody (4,7).
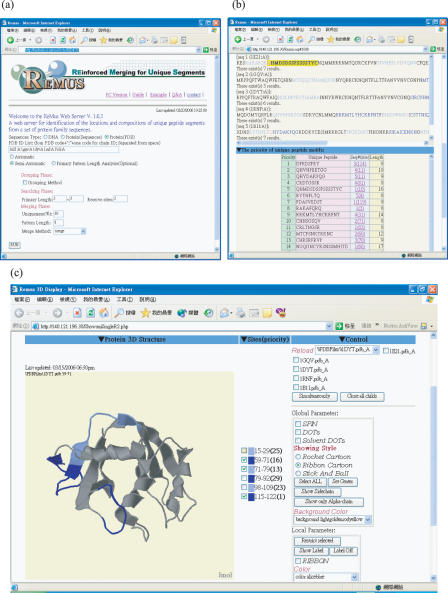
REMUS currently allows users to view the unique peptide segments in various kinds of three-dimensional images if there is structural information for the imported protein sequences. When the imported sequences are in PDB format, the system extracts the primary sequence information and identifies unique peptide segments in each sequence. Users are able to select unique peptide segments from priority lists to examine whether the segments are exposed to the surfaces at their elementary 3D structures. Figure 1a shows examples of REMUS web interface with five members of human RNaseA superfamily in PDB ID list (1e21:A, 1gqv:A, 1dyt:A, 1rnf:A and 1bli:A), Figure 1b depicts the results of identified unique peptide segments by automatic processing, and Figure 1c represents the positions of unique peptide segment in 3D visualization. The automatic mode of the REMUS system extracts unique peptide segments with a degree value above the default lower limit after performing the strict merging operation. The identified segments that fit in with the strict merging criterion with a length longer than the default limits are displayed in light or dark blue within their original sequences. The lengths are defined as 8, and the segments in purple indicate the overlapping residues located in the boundaries of two adjacent unique peptide segments. For the manual mode, a user has to assign various parameters based on his own need. The selective factors include substitution matrices, length of primary unique peptide, number of tolerant residues, length limit of merged peptide, merging methods and percentage limit of uniqueness levels. The final qualified unique peptide segments will be shown in various colors in order to distinguish individual segments. All the details and instructions are described in the system guidelines.
Figure 1.
Web interface of REMUS system. (a) The PDB IDs of five members of human RNaseA superfamily (1e21:A, 1gqv:A, 1dyt:A, 1rnf:A and 1b1i:A) are imported to REMUS. (b) The searched unique peptide segments for epitopes are displayed both in ranking order according to the antigenicity and highlighted in their original sequences, respectively. (c) The 3D structures of unique peptides with the first three highest priority for the 1dyt:A sequence are labeled in dark and light blue colors.
Evaluation of algorithm
To evaluate the performance of REMUS, the information related to epitopes of human monoclonal antibodies was retrieved from the website of Santa Cruz Biotechnology, Inc. (http://www.scbt.com/) which focused on the ongoing development of research antibodies. Among 8398 entries in the database, 83 monoclonal antibodies were generated against human antigens with specified epitopes <300 amino acid residues. These antigens were classified into 63 human protein families containing a total number of 264 protein sequences. Each set of the family protein sequences was collected from GenBank and analyzed by REMUS. It was found that 275 unique peptide segments were located within the antibody-antigen recognition sites of 66 monoclonal antibodies, indicating that the average accuracy of matching at least one of the unique peptide segments with the reported epitopes of the selected antibodies was 79.52% (66/83). As the lengths of the detected epitopes were decreased from 300, 200 to 100 amino acid residues, the accuracy of correlating a unique peptide segment with an epitope decreased from 94.12% (24/34), 81.25% (26/32), to 70.59% (16/17), respectively. Our results revealed that the accuracy was length-dependent and accuracy higher than 70% was successfully achieved.
DISCUSSION
REMUS has been applied for identification of unique sequential epitopes. The algorithm predicts potential epitopes based on the sequence features within a set of family protein sequences. To determine epitopes in a member of family for generation of peptide antigens or antibody drugs, several factors including hydrophilicity, antigenecity, surface accessibility and specificity have to be taken into consideration as most available web programs perform. In our REMUS program, the accumulation of non-gapped unique features is achieved by discriminating the common sequences to emphasize different regions from protein families. The REMUS system has been evaluated using several dataset including ribonuclease A, epidermal growth factor receptor, and matrix metalloproteinase protein families. In addition, the predicted unique peptide segments serving as sequential epitopes provide practical applications in designing experiments for specifically differentiating a member from a protein family or characterizing the antibody-recognition sites of protein antigens. The availability of the REMUS system for identifying unique peptide segments in protein families should be useful for further correlation between unique sequence motifs and specific protein functions.
Acknowledgments
This work is supported by National Science Council in Taiwan, Republic of China (NSC-93-3112-B-007-005, NSC94-2627-B-007-003 and NSC-94-3112-B-007-004-Y) and Veterans General Hospitals University System of Taiwan Joint Research Program (VGHUST94-G2–02-1). Funding to pay the Open Access publication charges for this article was provided by Center for Marine Bioscience and Biotechnology, National Taiwan Ocean University.
Conflict of interest statement. None declared.
REFERENCES
- 1.Zhang J., Dyer K.D., Rosenberg H.F. RNase8, a novel RNaseA superfamily ribonuclease expressed uniquely in placenta. Nucleic Acids Res. 2002;30:1169–1175. doi: 10.1093/nar/30.5.1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Houslay M.D., Schafer P., Zhang K.Y. Keynote review: phosphodiesterase-4 as a therapeutic target. Drug Discov. Today. 2005;10:1503–1519. doi: 10.1016/S1359-6446(05)03622-6. [DOI] [PubMed] [Google Scholar]
- 3.Rosenberg H.F., Dyer K.D. Eosinophil cationic protein and eosinophil-derived neurotoxin. Evolution of novel function in a primate ribonuclease gene family. J. Biol. Chem. 1995;270:30234. [PubMed] [Google Scholar]
- 4.Chang H.T., Pai T.W., Fan T.C., Su B.H., Wu P.C., Tang C.Y., Chang C.T., Liu S.H., Chang M.D.T. A reinforced merging methodology for mapping unique peptide motifs in members of protein families. BMC Bioinformatics. 2006;7:38. doi: 10.1186/1471-2105-7-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dayhoff M.O. A model of evolutionary change in proteins. Atlas Protein Seq. Struct. 1978;5:345–352. [Google Scholar]
- 6.Henikoff S., Henikoff J.G. Amino acid substitution matrices from protein blocks. Proc. Natl Acad. Sci. USA. 1992;89:10915–10919. doi: 10.1073/pnas.89.22.10915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boix E., Carreras E., Nikolovski Z., Cuchillo C.M., Nogues M.V. Identification and characterization of human eosinophil cationic protein by an epitope-specific antibody. J. Leukoc. Biol. 2001;69:1027–1035. [PubMed] [Google Scholar]