Abstract
We present POXO, a comprehensive tool series to discover transcription factor binding sites from co-expressed genes (www.bioinfo.biocenter.helsinki.fi/poxo). POXO manages tasks such as functional evaluation and grouping of genes, sequence retrieval, pattern discovery and pattern verification. It also allows users to tailor analytical pipelines from these tools, with single mouse clicks. One typical pipeline of POXO begins by examining the biological functions that a set of co-expressed genes are involved in. In this examination, the functional coherence of the gene set is evaluated and representative functions are associated with the gene set. This examination can also be used to group genes into functionally similar subsets, if several biological processes are affected in the experiment. The next step in the pipeline is then to discover over-represented nucleotide patterns from the upstream sequences of the selected gene sets. This enables to investigate the possibility that the genes are co-regulated by common cis-elements. If over-represented patterns are found, similar ones can then be clustered together and be verified. The performance of POXO is demonstrated by analysing expression data from pathogen treated Arabidopsis thaliana. In this example, POXO detected activated gene sets and suggested transcription factors responsible for their regulation.
INTRODUCTION
One of the central questions in biology is gene regulation: how and when are genes regulated and by what factors? In most cases, genes are regulated by transcription factors via cis-elements (1). In this regulatory mechanism, a transcription factor binds onto specific cis-elements that often are located in the first few thousand nucleotides upstream of the transcription start site (1). Thus, various genes that are located apart in the genome but work in concert can be simultaneously regulated by common transcription factors.
The regulatory circuits and the regulatory transcription factors of a gene or a set of genes can be investigated using computational methods. These computational methods are typically devised to discover over-represented nucleotide patterns that are then associated with the corresponding transcription factors. The use of these methods typically begins by gathering a set of putatively co-regulated genes. Next, gene functions can be examined, e.g. using gene ontology (GO) (2). GO terms offer a controlled vocabulary to describe the cellular component, molecular function and biological process of a gene (2). The statistical significance of the GO terms among the gathered gene set can be calculated and can be used to assign representative functions for a set of genes (3,4). If the gene set contains several distinct representative functions, it can be sensible to group the gene set into functionally coherent subsets (3,4). This can improve the signal-to-noise ratio of the binding site analysis, since genes with common functions also tend to share similar regulatory mechanisms (5,6). In particular, improvement can be expected, if the gene set contains genes involved in various different processes, each of which is controlled by its own cis-element.
Next, the upstream sequences of the selected genes are retrieved and potential cis-elements are searched for. In this binding site analysis, over-represented nucleotide patterns are considered as an evidence of cis-elements and co-regulation. Various tools exist for the task and they search for over-represented patterns using either probabilistic sequence models or pattern enumeration techniques (5–8). Probabilistic sequence models, like Gibbs samplers, discover long and general patterns (5,7). However, these tools do not always discover the most over-represented pattern, since they can get trapped in locally optimal solutions (6). Pattern enumerators analyze every pattern in the input set and therefore are always guaranteed to discover the most over-represented pattern, i.e. the globally optimal solution (6,8). However, pattern enumerators are restricted to relatively short pattern lengths, report multiple variants from a single original pattern and suffer from a limited pattern vocabulary.
After the discovery of over-represented nucleotide patterns, they should be verified and annotated. One approach to verify patterns is to visualize their positions within the sequences, since cis-elements are thought to locate at similar distances from the transcription start site (8). Another approach to verify patterns is to use evolutionary conservation among homologous sequences in different organisms (9,10). In this ‘phylogenetic footprinting’ approach homologous sequences are aligned. Since cis-elements are thought to remain unchanged and conserved during evolution, they should be found in the aligned regions (9,10). Patterns can also be annotated by screening collections of known binding sites (11). If a similar cis-element is found, it is possible that the query pattern acts as the matching cis-element and is recognized by the same transcription factor.
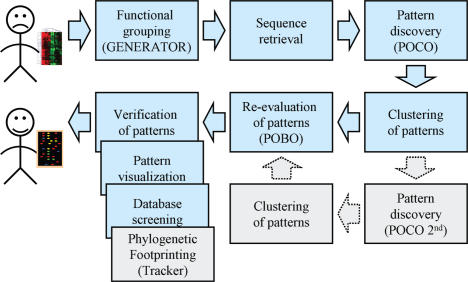
Here, we demonstrate POXO, a web-enabled tool series to discover transcription factors binding sites. POXO contains tools for various tasks that vary from functional evaluation and clustering of genes through sequence retrieval and pattern discovery to the evolutionary verification of patterns (Figure 1). These tools can be used independently or in pipelines, where the result of one tool is transferred to another one. Unlike currently existing tool series (12,13), POXO has also a tool to evaluate the coherence of the input gene set and to group these genes into functionally coherent subsets (4). The usability of POXO is demonstrated by analyzing a fungal pathogen treated Arabidopsis thaliana microarray experiment. In this example, POXO detected activated genes and suggested the transcription factors responsible for their regulation.
Figure 1.
Different tools in POXO. The computational pipeline and the tools used here to analyze the experimental data are highlighted by light blue arrows and boxes. Gray boxes and arrows indicate other available tools in POXO.
METHODS
The starting point is a long list of co-expressed genes from a biological analysis, e.g. genes expressed differentially between a treatment and its control. The individual tools integrated in POXO are briefly described below (Figure 1).
Stratifying a long gene list into functionally related subgroups
Functional sub-grouping enables the efficient analysis of large gene sets and enables to focus on genes involved in a specific function. Functional subsets can also improve the signal-to-noise ratio and highlight cis-elements that would have been otherwise missed, since genes involved in incoherent functions can mask the others' cis-elements. For example, in A.thaliana the attack of a pathogen can induce numerous genes (14). The number of genes varies from hundreds to thousands and the genes influenced by pathogens can be involved in notably different functions, such as response to biotic and abiotic stresses, metabolism and transport (14). Owing to the large amount of the induced genes and their dissimilar functions, it is unlikely that genes would be co-regulated by a single common transcription factor.
GENERATOR is a tool to evaluate the functional coherence of genes and to group the genes into functionally coherent subsets (4). In the tool, genes and their functional annotations from GO are represented as a binary matrix, which is factorized using non-negative matrix factorization (NMF) (15). NMF calculates an approximation (V ≈ WH) from an original matrix V (i × u) and creates two positive matrices H (a × u) and W (i × a), where a is the rank, i.e. the number of functionally coherent subsets created, i is the number of genes and u is the number of GO terms. NMF begins by randomly initializing the W and H matrices and it then iteratively updates the matrices using coupled divergence functions that minimize the least square error (LSE) of V and (WH) (15). When the algorithm stops, the resulting matrices (W and H) are used to resolve which subset each gene and term should be assigned to. GENERATOR assigns a gene to that subset which has the highest weight, i.e. a gene g is assigned to the subset s with the highest entry value wgs. Since NMF can terminate in locally optimal solutions, GENERATOR improves the reliability of the factorization by repeating the analysis several times and by representing only the best result, with respect to the calculated LSE.
Because it is difficult to predict the correct rank number before performing the analysis, GENERATOR automatically generates results using different ranks, i.e. it groups the genes into varying numbers of functionally coherent gene sets. The results of the different rank analyses are represented in a non-nested hierarchical tree, where the results of the analyses are visualized as bars. The bars are split into sections corresponding to the functional subsets. The width of each section corresponds to the number of genes belonging to the subset (4). The significance of the binary correlations between the subsets of different ranks is calculated. This enables to highlight those subsets of genes that are retained at different ranks (4). GENERATOR also represents the most representative GO term or function of the subset. These representative GO terms are selected according to their significance among the subset when compared to the transcriptome of the chosen organism using a hypergeometric distribution.
Sequence retrieval
The upstream sequences of the selected genes can be retrieved and further analyzed in POXO. POXO contains the upstream sequences of the known genes of several organisms: Anopheles gambiae, A.thaliana, Caenorhabditis elegans, Drosophila melanogaster, Homo sapiens, Mus musculus and Saccharomyces cerevisiae. The sequences are downloaded from TAIR (www.arabidopsis.org) (A.thaliana) or from Ensemble (www.ensembl.org) (16,17) and are recognized by their gene identifiers and accession numbers. The maximum sequence length is 3000 bp for all organisms except S.cerevisiae for which the maximum sequence length is 800 bp. The sequence collections are kept up-to-date and will be updated approximately twice a year.
Pattern discovery
Pattern discovery programs search for nucleotide patterns that are statistically significantly over-represented in a set of sequences in order to find patterns that are likely to share a regulatory function. POXO has a tool for pattern discovery called POCO (18). This same tool can also be used to search for patterns that maximize the discrimination between two sequence sets, being over-represented in one and under-represented in the other sequence set (18). The discriminative search can be helpful in situations where there are two oppositely behaving gene sets. For example, in A.thaliana different pathogens stimulate different defense responses and different signaling pathways, such as salicylic acid (SA), jasmonic acid (JA) and ethylene (E) mediated signaling pathways (14). While one pathogen activates genes involved in one pathway and represses genes involved in the others, a different pathogen activates and represses the gene sets conversely (14). This suggests that the gene sets could be regulated by different transcription factors and that the regulating cis-element of one gene set should not be the same as the regulating cis-elements of the other gene set. Thus, instead of searching over-represented nucleotide patterns from both sequence sets, it is possible to directly search discriminative patterns.
POCO is a pattern enumerator tool to discover statistically significant nucleotide patterns from either one or two sequence sets (18). In the tool, pattern distributions for each pattern in the input sequence sets and in the background sequence set are created using a bootstrap test. This enables the user to calculate the deviations for pattern occurrences and to use the deviation when assessing the statistical significance for patterns (18). If one sequence set is analyzed, patterns are evaluated using the t-test. If two sequence sets are analyzed, patterns are evaluated using ANOVA and are grouped into distinct pattern sets using Tukey's honestly significant post hoc test. The pattern sets are patterns over-represented in one of the sequence sets (pattern sets 1 and 2), patterns over-represented in both of the sequence sets (pattern set 3), and patterns over-represented in one and under-represented in the other sequence set (pattern sets 4 and 5) (18).
Discovery of co-occurring patterns
The possibility to search for patterns, which are over-represented in the vicinity of another pattern, enables to search for pattern combinations. These combinations could be, for example, cis-elements of two co-operating transcription factors, a long cis-element that was not discovered due to its length, or a cis-element that contains a variable length unspecific linker in the middle. For example, in A.thaliana some environmental stress inducible genes contain in their upstream regions two cis-elements, the dehydration response element (DRE) and the abscisic acid responsive element (ABRE) (19). These cis-elements are interdependent and both of them are needed for correct regulation (19).
‘POCO 2nd iteration’ is a tool to discover statistically significant nucleotide patterns that are located in the vicinity of a predetermined anchoring pattern, from either one or two sequence sets. In the tool, regions that are not in the vicinity of the anchoring pattern are masked and patterns are searched from the unmasked regions. Reliable background pattern distributions are created using only sequences that contain the anchoring pattern. The tool performs the analysis similarly to POCO and creates the same pattern sets (18). Since the same parameters can be used in this tool that were used in POCO, the obtained patterns and their test scores should be comparable between each other.
Clustering of patterns
Pattern enumerator tools, such as POCO, can report over-lapping patterns. For example, if pattern TTGACCG is over-represented, it is possible that also patterns such as TTGACC, TGACCG, TTNACC and the like are over-represented and reported in the analysis, since they can be derived from the same nucleotide pattern. To reduce the number of reported over-lapping patterns, similar patterns can be clustered together.
In POXO, patterns are clustered using the phi coefficient of correlation (φ) as distance (20) and hierarchical clustering. In clustering, the similarities of binary vectors describing the known nucleotide positions of the patterns are calculated (Equation 1) (20). The length of the binary vector corresponds to the total number of nucleotides within the sequences. Vector elements are coded as one if the nucleotide belongs to the pattern and as zero otherwise. In Equation 1, nTP indicates that the nucleotide belongs to both patterns, nFN and nFP indicate that the nucleotide only belongs to either one or the other of the patterns and nTN indicates that the nucleotide is not a part of either pattern.
| 1 |
In hierarchical clustering, the two closest vectors are iteratively merged together until a single vector remains. In iterations, two vectors are merged and distances are re-calculated. The new vector is the union of the merged vectors, i.e. it will contain all nucleotides of both patterns. The result of the phi coefficient of correlation ranges between −1 and 1, where −1 indicates mutually exclusive patterns, 1 indicates that the patterns are in identical positions and 0 indicates that the patterns are located randomly. In POXO, values that are <0 are reset to 0 to indicate random overlap. After clustering, a consensus pattern is created for the merged patterns by creating a pseudo-count corrected frequency weight matrix (21). The created consensus patterns can contain all possible IUPAC symbols. The statistical test scores for the clustered patterns can be easily calculated using POBO (see below), since the same parameters can be used in the evaluation that were used to create the original patterns.
Pattern evaluation
Often, a biologist will have a preliminary hypothesis about the regulatory factor and about its binding site. To quickly test the correctness of the hypothesis, POXO has a tool that can be used to screen and evaluate a predetermined pattern from either one or two sequence sets (22). In the POBO tool, pattern distributions are created similarly to POCO using a bootstrap test, after which the significance of the pattern is evaluated using the t-test or ANOVA. In POBO, the predetermined pattern can be represented using IUPAC codes, regular expressions or pattern weight matrices. Patterns can also be screened allowing mismatches.
Pattern visualization
POXO has a tool to visualize the locations of the discovered patterns. This can be used to verify patterns and to examine their locations in the sequences. The visualization illustrates if patterns are located approximately at the same distance from the transcription start site, as is assumed for cis-elements. The visualization can also illustrate if two patterns frequently co-occur together.
Phylogenetic footprinting
Patterns can also be verified using phylogenetic footprinting, in which the conservation of the patterns is examined in homologous sequences across different organisms. In this method functionally important cis-elements are assumed to remain unchanged during evolution and to be located in the conserved regions in different organisms. This method has been shown to decrease false positive predictions (9,10).
Tracker is a tool to analyze the evolutionary conservation of the discovered patterns in homologous sequences and it can be used for the previously mentioned organisms except A.thaliana. In the tool all homologous gene relationships between the genes on different organisms are gathered from Ensemble (www.ensembl.org) (17).
In Tracker, query genes and their corresponding upstream sequences can be retrieved using either Ensemble ids (gene identifiers) or Blast searches (23). If Blast searches are used, then the query sequences, e.g. sequences used in the binding site analysis, are first blasted against the stored upstream sequences in POXO. If a plausible match is found, the homologous gene relationships of the gene associated to the matching upstream sequence are used. After exploring the query genes and the inferred homologous genes on other organisms, their upstream sequences are retrieved. Next, the pair-wise alignments between the query sequence and all its homologous sequences are created using PromoterWise (24). These pair-wise alignments are then assembled together and, to provide an easy visual interpretation of the results, gaps in the query sequence are closed by removing insertions in the homologous sequences. After the alignment is done, the discovered patterns are screened to visualize their locations within the alignment. Evolutionary conserved patterns should occur in the aligned regions and be located at the same vertical positions relative to the query sequence.
Database screening
Patterns can be annotated by screening for similar cis-elements from collections of known cis-elements (11). If matching cis-elements and associated transcription factors are found, it can be speculated that also the discovered patterns are binding the same transcription factors as their matches. In POXO, discovered patterns can be annotated by screening either a plant specific or more general cis-elements collections, retrieved from PLACE, TRANSFAC (public) and JASPAR (25–27). In the pattern screens, patterns of similar length and patterns having at least a certain amount of similar nucleotides are reported.
Availability and running the program
All tools in POXO are written using Perl, C or C++. A MySQL (www.mysql.com) database is used to store the background sequences and gene relationships. In POCO and POCO 2nd iteration, P-values are calculated using the DCDFLIB-package (www.netlib.org/random). The server, help-pages, background models for some tools (MySQL dump-files) and the source codes of the tools are available from the group's web page (www.bioinfo.biocenter.helsinki.fi/poxo). Some tools require Adobe SVG Viewer (www.adobe.com/svg/), which is a freeware browser plug-in to view scalable vector graphics.
Experimental data acquisition and analysis
For data acquisition and for parameters used see Supplementary Data.
RESULTS AND DISCUSSION
To demonstrate how to use POXO, gene expression data from a Botrytis cinerea treated A.thaliana microarray experiment was re-analyzed using the pipeline depicted in Figure 1. B.cinerea is a necrotrophic fungal pathogen of A.thaliana. Resistance against the pathogen involves co-operation of various defense pathways, but ethylene (E) and salicylic acid (SA) pathways are though to orchestrate the defense response (28). In the experiment, the gene expression of B.cinerea infected A.thaliana was measured 18 and 48 h after the infection (28). Here, the gene expression data were re-analyzed and clustered into gene sets that contained genes activated during the infection (1222 genes) and genes repressed during the infection (1626 genes).
Results on the experimental data using POXO
The first step in the pipeline is to group the genes into functionally coherent and more specific subsets. The grouping revealed that both clusters contained several functionally distinct subsets (Supplementary Tables 1 and 2). One of the top subsets in both clusters was enriched in genes involved in response to stimulus (GO:0050896). Among the activated genes, the response to stimulus subset contained 139 genes at rank 5. Other interesting and enriched functions among this subset were defense response (GO:0006952), response to pest, pathogen or parasite (GO:0009613), JA and E-dependent systemic resistance (GO:0009861) and response to chitin (GO:0010200). (Supplementary Table 3). Among the repressed genes, the response to stimulus subset contained 144 genes at rank 1. Other enriched functions were response to abiotic stimulus (GO:0009628) and response to auxin stimulus (GO:0009733) (Supplementary Table 4).
The next step in the pipeline is to retrieve the upstream sequences of the subsets and to discover over-represented nucleotide patterns, which could explain the observed co-expression. In binding site analysis, the top 10 patterns over-represented in one and under-represented in the other sequence set were searched (Supplementary Tables 5 and 6). Next, these 10 patterns were clustered together (Supplementary Figures 1 and 2).
Pattern clustering produced seven distinct patterns for the activated subset, i.e. patterns over-represented among the activated and under-represented among the repressed subset (Table 1). To annotate the patterns, similar cis-elements were screened from the collections of known plant binding sites (25). Screening yielded two cis-elements, which had a biologically meaningful association to the observed functions. CTGAGGAA resembled the cis-element of NPR1 (CTGAAGAAGAA, matching part bold and underlined) (29), which is the key transcription factor in the SA pathway. It is also believed that NPR1 is involved in resistance to B.cinerea (28). GGAAAANG resembled a cis-element (GAAAAA) that plays a role in pathogen and salt induced gene expression in Glycine max (30). (The locations of the patterns are shown in Supplementary Figure 3.) For the other subset, i.e. patterns over-represented among the repressed and under-represented among the activated subset, six patterns were generated (Table 2). When these patterns were screened against the cis-element collection, two known auxin responsive elements in G.max were found (TTTTGTCAC resembling CCTTTTGTCTC and GGACCNA resembling GGTCCCAT) (31,32). (The locations of the patterns are in Supplementary Figure 4.)
Table 1.
The seven clustered patterns over-represented in the activated response to stimulus (GO:0050896) gene set and under-represented in the repressed response to stimulus gene set
| Pattern | Activated genes | Repressed genes | P-value | min P | Ac | ||
|---|---|---|---|---|---|---|---|
| occ | pro | occ | pro | ||||
| TGGAAd/TTCCA | 651 | 139 | 508 | 139 | 5E−05 | 5E−05 | S000403 |
| GGAAAANG/CNTTTTCC | 62 | 50 | 23 | 23 | 2E−04 | 2E−04 | S000453 |
| CATNNCGG/CCGNNATG | 36 | 34 | 9 | 9 | 3E−04 | 3E−04 | S000250 |
| TGCGANNC/GNNTCGCA | 38 | 36 | 11 | 11 | 4E−04 | 4E−04 | |
| GTVATCCT/AGGATBAC | 26 | 23 | 5 | 4 | 1E−03 | 1E−04 | S000470 |
| TNCNAGG/CCTNGNA | 200 | 100 | 127 | 86 | 7E−04 | 7E−04 | |
| CTGAGGAA/TTCCTCAG | 14 | 14 | 1 | 1 | 3E−03 | 3E−04 | S000473 |
In the table occ is the pattern occurrence, pro is the number of promoters with the pattern, P-value is the significance of the clustered pattern, min P is the minimum P-value of the original patterns used in clustering and Ac is the accession code of the best match found in PLACE (25).
Table 2.
The six clustered patterns under-represented in the activated response to stimulus gene set and over-represented in the repressed response to stimulus gene set
| Pattern | Activated genes | Repressed genes | P-value | min P | Ac | ||
|---|---|---|---|---|---|---|---|
| occ | pro | occ | pro | ||||
| TNGGTCC/GGACCNA | 34 | 27 | 79 | 62 | 4E−04 | 4E−04 | S000360 |
| CTTTGCNT/ANGCAAAG | 21 | 19 | 64 | 47 | 5E−04 | 5E−04 | S000354 |
| GNANTATA/TATANTNC | 144 | 84 | 229 | 107 | 5E−04 | 5E−04 | |
| TGTGATTGG/CCAATCACA | 3 | 3 | 14 | 13 | 1E−02 | 1E−04 | S000143 |
| CAWTKATTG/CAATMAWTG | 18 | 18 | 26 | 23 | 5E−01 | 3E−04 | S000371 |
| TTTTGTCAC/GTGACAAAA | 3 | 3 | 5 | 6 | 1E+00 | 7E−05 | S000337 |
Notation as in Table 1.
CONCLUSION
We have developed a tool series called POXO, which is accessible via a web interface. POXO can be used to discover putative transcription factor binding sites from a set of genes. We have also demonstrated, by example, how these different tools can be used to discover biologically meaningful binding sites from the vast amount of data.
In the example, a pathogen treated A.thaliana experiment was analyzed using POXO. In this analysis, two functionally interesting subsets were detected. The first functional subset contained 139 genes, which were activated during the infection and which were involved in the response to stimulus function. Some of the genes within this subset are involved in pathogen defense. Interestingly, the binding site analysis discovered over-represented nucleotide patterns that resemble known cis-elements associated with SA-mediated defense pathway and pathogen defense (29,30). The other functionally interesting gene set contained 144 genes, which were repressed during the infection and which GO annotates with the response to stimulus function. Some of these genes were involved in the auxin stimulus and, interestingly, two over-represented patterns among the upstream sequences of these genes were discovered to resemble known auxin responsive elements (31,32). Thus, for both functional subsets, POXO was successfully used to find the regulated genes as well as potential transcription factors regulating them.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
Acknowledgments
We thank Christopher Wilton and Günter Brader for helpful discussions about the manuscript. This work was supported by the Academy of Finland (grant no. 1105182) and by a grant from the Ministry of Education to M.K. P.P. would like to thank the Finnish Cultural Foundation for financial support. Funding to pay the Open Access publication charges for this article was provided by the Academy of Finland.
Conflict of interest statement. None declared
REFERENCES
- 1.Wray G.A., Hahn M.W., Abouheif E., Balhoff J.P., Pizer M., Rockman M.V., Romano L.A. The evolution of transcriptional regulation in eukaryotes. Mol. Biol. Evol. 2003;20:1377–1419. doi: 10.1093/molbev/msg140. [DOI] [PubMed] [Google Scholar]
- 2.Ashburner M., Ball C.A., Blake J.A., Botstein D., Butler H., Cherry J.M., Davis A.P., Dolinski K., Dwight S.S., Eppig J.T., et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nature Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beissbarth T., Speed T.P. GOstat: find statistically overrepresented gene ontologies within a group of genes. Bioinformatics. 2004;20:1464–1465. doi: 10.1093/bioinformatics/bth088. [DOI] [PubMed] [Google Scholar]
- 4.Pehkonen P., Wong G., Toronen P. Theme discovery from gene lists for identification and viewing of multiple functional groups. BMC Bioinformatics. 2005;6:162. doi: 10.1186/1471-2105-6-162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hughes J.D., Estep P.W., Tavazoie S., Church G.M. Computational identification of cis-regulatory elements associated with groups of functionally related genes in Saccharomyces cerevisiae. J. Mol. Biol. 2000;296:1205–1214. doi: 10.1006/jmbi.2000.3519. [DOI] [PubMed] [Google Scholar]
- 6.Sinha S., Tompa M. Discovery of novel transcription factor binding sites by statistical overrepresentation. Nucleic Acids Res. 2002;30:5549–5560. doi: 10.1093/nar/gkf669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Thijs G., Lescot M., Marchal K., Rombauts S., De Moor B., Rouzé P., Moreau Y. A higher order background model improves the detection of regulatory elements by Gibbs sampling. Bioinformatics. 2001;17:1113–1122. doi: 10.1093/bioinformatics/17.12.1113. [DOI] [PubMed] [Google Scholar]
- 8.van Helden J., André B., Collado-Vides J. Extracting regulatory sites from the upstream region of yeast genes by computational analysis of oligonucleotide frequencies. J. Mol. Biol. 1998;281:827–842. doi: 10.1006/jmbi.1998.1947. [DOI] [PubMed] [Google Scholar]
- 9.Ho Sui S.J., Mortimer J.R., Arenillas D.J., Brumm J., Walsh C.J., Kennedy B.P., Wasserman W.W. oPOSSUM: identification of over-represented transcription factor binding sites in co expressed genes. Nucleic Acids Res. 2005;33:3154–3164. doi: 10.1093/nar/gki624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Boffelli D., McAuliffe J., Ovcharenko D., Lewis K.D., Ovcharenko I., Pachter L., Rubin E.M. Phylogenetic shadowing of primate sequences to find functional regions of the human genome. Science. 2003;299:1391–1394. doi: 10.1126/science.1081331. [DOI] [PubMed] [Google Scholar]
- 11.Roepcke S., Grossmann S., Rahmann S., Vingron M. T-Reg comparator: an analysis tool for the comparison of position weight matrices. Nucleic Acids Res. 2005;33:W438–W441. doi: 10.1093/nar/gki590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.van helden J. Regulatory sequence analysis tools. Nucleic Acids Res. 2003;31:3593–3596. doi: 10.1093/nar/gkg567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Aerts S, Thijs G., Coessens B., Staes M., Moreau Y., De Moor B. Toucan: deciphering the cis-regulatory logic of coregulated genes. Nucleic Acids Res. 2003;31:1753–1764. doi: 10.1093/nar/gkg268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.De Vos M., Van Oosten V.R., Van Poecke R.M., Van Pelt J.A., Pozo M.J., Mueller M.J., Buchala A.J., Metraux J.P., Van Loon L.C., Dicke M., et al. Signal signature and transcriptome changes of Arabidopsis during pathogen and insect attack. Mol. Plant. Microbe. Interact. 2005;18:923–937. doi: 10.1094/MPMI-18-0923. [DOI] [PubMed] [Google Scholar]
- 15.Lee D.D., Seung H.S. Learning the parts of objects by non-negative matrix factorization. Nature. 1999;401:788–791. doi: 10.1038/44565. [DOI] [PubMed] [Google Scholar]
- 16.Rhee S.Y., dBeavis W., Berardini T.Z., Chen G., Dixon D., Doyle A., Garcia-Hernandez M., Huala E., Lander G., Montoya M., et al. The Arabidopsis Information Resource (TAIR): a model organism database providing a centralized, curated gateway to Arabidopsis biology, research materials and community. Nucleic Acids Res. 2003;31:224–228. doi: 10.1093/nar/gkg076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Birney E., Andrews D., Caccamo M., Chen Y., Clarke L., Coates G., Cox T., Cunningham F., Curwen V., Cutts T., et al. Ensembl 2006. Nucleic Acids Res. 2006;34:D556–D561. doi: 10.1093/nar/gkj133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kankainen M., Holm L. POCO: discovery of regulatory patterns from promoters of oppositely expressed gene sets. Nucleic Acids Res. 2005;33:W427–W431. doi: 10.1093/nar/gki467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Narusaka Y., Nakashima K., Shinwari Z.K., Sakuma Y., Furihata T., Abe H., Narusaka M., Shinozaki K., Yamaguchi-Shinozaki K. Interaction between two cis-acting elements, ABRE and DRE, in ABA-dependent expression of Arabidopsis rd29A gene in response to dehydration and high-salinity stresses. Plant J. 2003;34:137–148. doi: 10.1046/j.1365-313x.2003.01708.x. [DOI] [PubMed] [Google Scholar]
- 20.Burset M., Guigo R. Evaluation of gene structure prediction programs. Genomics. 1996;34:353–367. doi: 10.1006/geno.1996.0298. [DOI] [PubMed] [Google Scholar]
- 21.Hertz G.Z., Stormo G.D. Identifying DNA and protein patterns with statistically significant alignments of multiple sequences. Bioinformatics. 1999;15:563–577. doi: 10.1093/bioinformatics/15.7.563. [DOI] [PubMed] [Google Scholar]
- 22.Kankainen M., Holm L. POBO, transcription factor binding site verification with bootstrapping. Nucleic Acids Res. 2004;32:W222–W229. doi: 10.1093/nar/gkh463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 24.Ettwiller L., Paten B., Souren M., Loosli F., Wittbrodt J., Birney E. The discovery, positioning and verification of a set of transcription-associated motifs in vertebrates. Genome Biol. 2005;6:R104. doi: 10.1186/gb-2005-6-12-r104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Higo K., Ugawa Y., Iwamoto M., Korenaga T. Plant cis-acting regulatory DNA elements (PLACE) database:1999. Nucleic Acids Res. 1999;27:297–300. doi: 10.1093/nar/27.1.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Matys V., Kel-Margoulis O.V., Fricke E., Liebich I., Land S., Barre-Dirrie A., Reuter I., Chekmenev D., Krull M., Hornischer K., et al. TRANSFAC and its module TRANSCompel: transcriptional gene regulation in eukaryotes. Nucleic Acids Res. 2006;34:D108–D110. doi: 10.1093/nar/gkj143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vlieghe D., Sandelin A., De Bleser P.J., Vleminckx K., Wasserman W.W., van Roy F., Lenhard B. A new generation of JASPAR, the open-access repository for transcription factor binding site profiles. Nucleic Acids Res. 2006;34:D95–D97. doi: 10.1093/nar/gkj115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ferrari S., Plotnikova J.M., De Lorenzo G., Ausubel F.M. Arabidopsis local resistance to Botrytis cinerea involves salicylic acid and camalexin and requires EDS4 and PAD2, but not SID2, EDS5 or PAD4. Plant J. 2003;35:193–205. doi: 10.1046/j.1365-313x.2003.01794.x. [DOI] [PubMed] [Google Scholar]
- 29.Wang D., Weaver N.D., Kesarwani M., Dong X. Induction of protein secretory pathway is required for systemic acquired resistance. Science. 2005;308:1036–1040. doi: 10.1126/science.1108791. [DOI] [PubMed] [Google Scholar]
- 30.Park H.C., Kim M.L., Kang Y.H., Jeon J.M., Yoo J.H., Kim M.C., Park C.Y., Jeong J.C., Moon B.C., Lee J.H., et al. Pathogen- and NaCl-induced expression of the SCaM-4 promoter is mediated in part by a GT-1 box that interacts with a GT-1-like transcription factor. Plant Physiol. 2004;135:2150–2161. doi: 10.1104/pp.104.041442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ulmasov T., Murfett J., Hagen G., Guilfoyle T.J. Aux/IAA proteins repress expression of reporter genes containing natural and highly active synthetic auxin response elements. Plant Cell. 1997;9:1963–1971. doi: 10.1105/tpc.9.11.1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xu N., Hagen G., Guilfoyle T. Multiple auxin response modules in the soybean SAUR 15A promoter. Plant Sci. 1997;126:193–201. [Google Scholar]