Abstract
Obtaining high quality multiple alignments is crucial for a range of sequence analysis tasks. A common strategy is to align the sequences several times, varying the program or parameters until the best alignment according to manual inspection by human experts is found. Ideally, this should be assisted by an automatic assessment of the alignment quality. Our web-site http://msa.cgb.ki.se allows users to perform all these steps: Kalign to align sequences, Kalignvu to view and verify the resulting alignments and Mumsa to assess the quality. Due to the computational efficiency of Kalign we can allow users to submit hundreds of sequences to be aligned and still guarantee fast response times. All servers are freely accessible and the underlying software can be freely downloaded for local use.
INTRODUCTION
High-quality multiple alignments are required for many sequence analysis tasks, e.g. homology detection using profiles, evolutionary tree reconstruction and function/structure analysis of protein families (1,2). Although the field of multiple alignment has received a lot of attention recently (3–5) several issues remain. A central problem is that alignment programs are sensitive to parameter settings, such as gap penalties. Conversely, different protein families often require different sets of parameters. It is therefore unlikely that using default parameters, usually obtained via training on small benchmark sets, would give accurate alignments for an individual alignment case.
One way to overcome this problem is to repeat the alignment procedure several times with different parameters until a good alignment is obtained. This interactive process can also reveal biological relationships between the sequences that are not readily visible from single alignments. For example, an alignment obtained with low gap penalties shows which sequences are close relatives, because only these sequences will be aligned, while an alignment with stringent penalties will highlight conserved blocks that all sequences share. Moreover, each step or alignment in the interactive process can enhance the understanding of the evolutionary relationships among the analyzed sequences. In our experience alignments obtained in such a way are consistently more accurate than alignments obtained by running any given alignment program just once. However, visual detection of ‘incorrect’ multiple alignments can be challenging, and errors in the alignment may go undetected. It is therefore of great value to have a tool that performs the comparison of several multiple alignments automatically in order to identify potentially incorrect alignments. We here present an online suite of easy-to-use servers that allow users to perform all steps necessary to arrive at high-quality alignments. This includes the fast sequence alignment program Kalign, the lightweight alignment viewer Kalignvu, and the alignment accuracy assessment program Mumsa.
SERVERS
Kalign
Our alignment program Kalign (6) is among the fastest and best performing alignment algorithms available. A key feature is its computational performance, which is very important for the interactive alignment strategy outlined above. On average Kalign takes less than a second to align one hundred protein sequences of length 500.
The server version of Kalign can be used with a subset of the most important parameters available in the command line version. These options include three different types of gap penalties: the gap open, internal extension and terminal extension penalty. It is also possible to add a bonus score to all fields of the substitution matrix. Although Kalign can align both protein and nucleotide sequences we here limit our discussion to the alignment of protein sequences.
The gap open and internal extension penalties are the standard penalties employed in all alignment programs using the affine gap model. In Kalign, half of the gap open penalty is applied to the start and half to the end of a gap. This ensures that both borders near gaps are treated equally. The internal extension penalty is applied for each elongation of a gap within a sequence. Similarly, the terminal extension penalty is applied just as the extension penalty but to N or C-terminal gaps. Due to a quirk in our dynamic programming implementation, half of the gap open penalty is also applied when leaving an N-terminal gap and upon entering a C-terminal gap. The bonus score parameter, unique to Kalign, deserves some special attention. Some alignment programs use all-positive matrices, which aids in the alignment of remote homologs (7). However, using all-positive matrices can have the undesirable effect of aligning non-homologous sequences to each other. To balance these two aspects we decided to allow users to specify a parameter to be added to all fields of the substitution matrix. Set to zero, the default substitution matrix [Gonnet (8) for proteins and HOXD (9) for nucleotides] is used; set to the absolute value of the lowest substitution score (5.2 for proteins and 125 for nucleotides), the matrix becomes all-positive.
The recommended strategy for obtaining good alignments with Kalign is to start with less stringent parameters (the default on our web-site) and gradually increase the gap open and internal extension penalties. Care has to be taken when using the bonus score and terminal gap penalty as these parameters can force Kalign to align non-homologous sequences. The optimal settings for the terminal extension penalty depend on whether full-length or fragmented sequences (e.g. variable domains) are present. In the former case a good range is between zero and about half the internal extension penalty (10). With fragmented sequences the terminal extension penalty should be set to equal the internal penalty. In general, we recommend using a low bonus score (maximum 0.2) and a higher one only in cases where sequences are known to be homologous over the entire length. All of these recommendations are of course not accurate for all alignment cases and we encourage users to experiment with the parameters.
Kalignvu
The visual inspection of alignments is a crucial step in ensuring alignment quality. To facilitate this process online we designed an xml-based alignment viewer: Kalignvu. Key features include its ability to display sequence names during horizontal scrolling, the option to resize the alignment and support for different colour schemes or types of sequence annotation. Since Kalignvu is xml-based, resizing the alignment or choosing different colour schemes does not require resubmitting information to a server or reloading of the entire page. This makes Kalignvu quick and responsive.
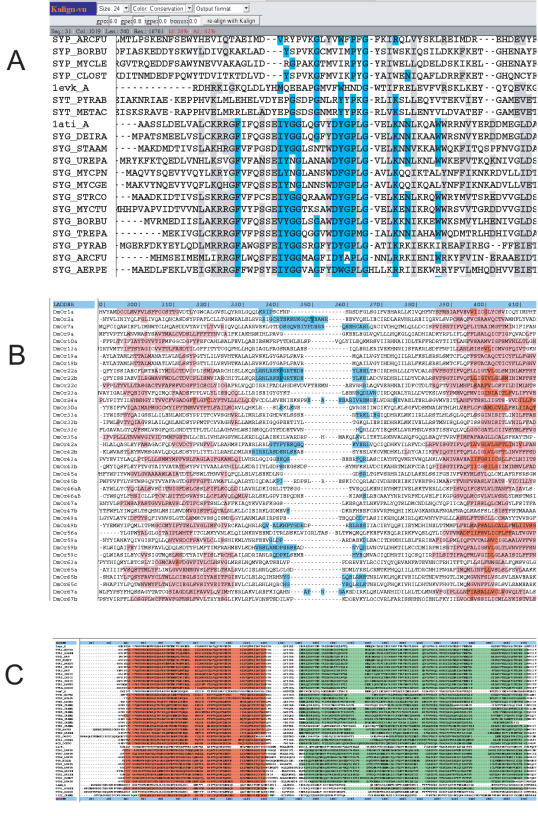
For protein sequences Kalignvu offers three colour schemes adopted from Belvu (http://www.cgb.ki.se/cgb/groups/sonnhammer/Belvu.html): two based on residue type and one based on conservation (Figure 1A). In addition, there are two hydrophobicity schemes available, calculated by the Kyte–Doolittle method (11) at window lengths of 7 and 21. The former can be used to differentiate between buried and surface residues in globular proteins while the latter is more suited to identify potential transmembrane regions (Figure 1B). Three nucleotide colour schemes are also available (two based on residue type and one based on conservation). Finally, using the Macsim format (see supported input/output formats) Kalignvu can display any user-provided features associated with sequences, such as secondary structure, alternative splice sites or Pfam domains (Figure 1C).
Figure 1.
Alignments viewed by Kalignvu with different color schemes and font sizes. (A) Balibase reference alignment ‘BB30002’ with the default conservation scheme. (B) An alignment of odorant receptors colored by the hydrophobicity colour scheme (window length of 21). Kalignvu implements a scale from blue to red in nine steps corresponding to the minimum–maximum hydrophobicity. Here, the red segments indicate transmembrane domains and the blue segments an interconnecting hydrophilic loop. Note that most gaps occur within the hydrophilic loops. (C) Balibase reference alignment ‘BB30014’ with Pfam domains ‘PF00043’ and ‘PF02798’ highlighted.
To fully integrate the viewer into our site, Kalignvu allows users to directly use Kalign to realign the sequences with a new set of parameters. This feature is essential for the interactive alignment strategy central to our site.
To aid in determining alignment quality the average percentage identity and percentage of aligned residues for each alignment are given. The latter is the fraction of aligned residues divided by the total theoretical number of possible aligned residues. An increase in both these values usually indicates that the current alignment is of higher quality than the previous one. Once an adequate alignment is achieved, Kalignvu allows users to download the alignments in a variety of formats.
Mumsa
Mumsa (12) is a tool for automatic assessment of alignment quality. To use the Mumsa server, a number of alternate multiple alignments have to be generated and submitted. The server then computes the average overlap score (AOS), reflecting the difficulty of aligning the sequences and a multiple overlap score (MOS) indicating the quality of each individual alignment. Both scores range between one and zero.
The AOS score is very important in determining how trustworthy an alignment can be expected to be. If the alignment is used for further purposes, such as phylogenetic tree reconstruction, knowing the quality of the underlying alignment is of great value. We here provide some general rules of thumb for quality assessment. An AOS score above 0.8 indicates good agreement among the input alignments (12), meaning that the sequences are easy to align and the alignments can probably be trusted. However, if the AOS score drops below 0.5 the sequences are very difficult to align and the respective input alignments have to be treated with care.
The MOS score can be used for picking the best alignment among alternate solutions. As a rule of thumb, alignments with a MOS score above 0.8 may be considered reliable. Both scores are important and have to be considered jointly. For example, the best alignment according to the MOS score is probably not accurate enough for further studies if the AOS score for the entire alignment case is very low. For large-scale projects we recommend using a local copy of Mumsa.
Supported input/output formats
Both Kalign and Kalignvu support the following multiple sequence alignment formats for both input and output: aligned Fasta, Stockholm (http://www.cgb.ki.se/cgb/groups/sonnhammer/Stockholm.html) MSF, Clustal and the Macsim xml format used for the Balibase 3.0 (13) database. Kalignvu can therefore be used to convert alignments from one format into another. In addition, Kalign accepts unaligned input sequences in Fasta, Uniprot flatfile or xml format.
At the moment, Mumsa requires all alignments to be in aligned Fasta format and have the sequences in the same order.
Implementation/availability
All three servers present on our site can be downloaded in the form of freely available stand-alone C programs. Kalignvu is particularly useful for bioinformatics servers displaying alignments, such as Pfam (14) and Funshift (15).
CONCLUSIONS
Our centralized site contains several servers that cover all the steps necessary to obtain high quality alignments. A common interface to our servers means they are easy to use, and practical problems concerning different input/output formats are avoided. In response to the demand of performing ever larger alignments, fueled by the increase of data from sequencing projects, our servers allow users to submit hundreds of sequences at a time. To our knowledge this service is unique.
In conclusion, we provide the community with a powerful, yet easy to use suite of tools for multiple sequence alignment.
Acknowledgments
The authors would like to thank Lukas Käll and Abhiman Saraswathi for many useful discussions and Bent Terp for maintaining the physical server. Funding to pay the Open Access publication charges for this article was provided by Swedish Graduate School for Functional Genomics and Bioinformatics.
Conflict of interest statement. None declared.
REFERENCES
- 1.Lecompte O., Thompson J.D., Plewniak F., Thierry J., Poch O. Multiple alignment of complete sequences (MACS) in the postgenomic era. Gene. 2001;270:17–30. doi: 10.1016/s0378-1119(01)00461-9. [DOI] [PubMed] [Google Scholar]
- 2.Notredame C. Recent progress in multiple sequence alignment: a survey. Pharmacogenomics. 2002;3:131–144. doi: 10.1517/14622416.3.1.131. [DOI] [PubMed] [Google Scholar]
- 3.Do C.B., Mahabhashyam M.S.P., Brudno M., Batzoglou S. ProbCons: probabilistic consistency-based multiple sequence alignment. Genome Res. 2005;15:330–340. doi: 10.1101/gr.2821705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Edgar R.C. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Katoh K., Kuma K., Toh H., Miyata T. MAFFT version 5: improvement in accuracy of multiple sequence alignment. Nucleic Acids Res. 2005;33:511–518. doi: 10.1093/nar/gki198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lassmann T., Sonnhammer E.L.L. Kalign–an accurate and fast multiple sequence alignment algorithm. BMC Bioinformatics. 2005;6:298. doi: 10.1186/1471-2105-6-298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Thompson J.D., Higgins D.G., Gibson T.J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gonnet G.H., Cohen M.A., Benner S.A. Exhaustive matching of the entire protein sequence database. Science. 1992;256:1443–1445. doi: 10.1126/science.1604319. [DOI] [PubMed] [Google Scholar]
- 9.Chiaromonte F., Yap V.B., Miller W. Proceedings of the Pacific Symposium on Biocomputing. 2002. Scoring pairwise genomic sequence alignments; pp. 115–126. [DOI] [PubMed] [Google Scholar]
- 10.Edgar R.C. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics. 2004;5:113. doi: 10.1186/1471-2105-5-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kyte J., Doolittle R.F. A simple method for displaying the hydropathic character of a protein. J. Mol. Biol. 1982;157:105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- 12.Lassmann T., Sonnhammer E.L.L. Automatic assessment of alignment quality. Nucleic Acids Res. 2005;33:7120–7128. doi: 10.1093/nar/gki1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Thompson J.D., Koehl P., Ripp R., Poch O. BaliBASE 3.0: Latest developments of the multiple sequence alignment benchmark. Proteins. 2005;61:127–136. doi: 10.1002/prot.20527. [DOI] [PubMed] [Google Scholar]
- 14.Finn R.D., Mistry J., Schuster-Bockler B., Griffiths-Jones S., Hollich V., Lassmann T., Moxon S., Marshall M., Khanna A., Durbin R., et al. Pfam: clans, web tools and services. Nucleic Acids Res. 2006;34:D247–D251. doi: 10.1093/nar/gkj149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Abhiman S., Sonnhammer E.L.L. FunShift: a database of function shift analysis on protein subfamilies. Nucleic Acids Res. 2005;33:D197–D200. doi: 10.1093/nar/gki067. [DOI] [PMC free article] [PubMed] [Google Scholar]