Abstract
ProMiR is a web-based service for the prediction of potential microRNAs (miRNAs) in a query sequence of 60–150 nt, using a probabilistic colearning model. Identification of miRNAs requires a computational method to predict clustered and nonclustered, conserved and nonconserved miRNAs in various species. Here we present an improved version of ProMiR for identifying new clusters near known or unknown miRNAs. This new version, ProMiR II, integrates additional evidence, such as free energy data, G/C ratio, conservation score and entropy of candidate sequences, for more controllable prediction of miRNAs in mouse and human genomes. It also provides a wider range of services, e.g. the prediction of miRNA genes in long nonrelated sequences such as viral genomes. Importantly, we have validated this method using several case studies. All data used in ProMiR II are structured in the MySQL database for efficient analysis. The ProMiR II web server is available at http://cbit.snu.ac.kr/~ProMiR2/.
INTRODUCTION
MicroRNAs (miRNAs) constitute a large family of noncoding RNAs, which take part directly in posttranscriptional regulation either by arresting the translation of mRNAs or by their cleavage (1). miRNAs are defined as single-stranded RNAs of ∼22 nt in length (range 19–25 nt) generated from endogenous transcripts that can form local hairpin structures (2).
Since the discovery of lin-4 and let-7, efforts to identify miRNA genes have led to the discovery of hundreds of miRNAs in animals, plants and viruses (3–6). All of them have been archived in miRBase (http://microrna.sanger.ac.uk/sequences/). High-throughput miRNA identification has been accomplished by directional cloning of endogenous small RNAs (7,8). However, a limitation of this approach is that miRNAs expressed at low levels or only in a specific condition or specific cell types are difficult to detect.
Computational approaches can overcome this problem, at least in part. They are based on the structural and sequential characteristics of miRNA precursors. Previous computational approaches for miRNA prediction have mainly searched for miRNAs that are closely homologous to published miRNAs (9–11). However, such methods failed to detect any new families that lacked clear homologues. In particular, several miRNAs with genus-specific patterns require a method to predict unrelated miRNA genes. Several approaches have been proposed to search for new miRNA families using comparative genomics, based on regulatory motifs in conserved DNA and with patterns conserved among the sequences and structures of previously studied distant families (12–14).
ProMiR has been used successfully to predict an miRNA in a stem–loop sequence using a score generated by a probabilistic colearning model without any other evidence (15). Here we introduce an improved method to identify the conserved and nonconserved miRNAs near known miRNAs or candidates. This strategy is very useful because more than half of the known miRNA genes are present as tandem arrays within operon-like clusters. This new version, ProMiR II, generates a list of nearby potential miRNAs according to score and to several filtering criteria such as conservation score, entropy, G/C ratio and free energy. This enhanced method allows for low- or high-stringency prediction of conserved and nonconserved miRNA genes by adjusting the filtering criteria. Importantly, we have used it to validate the prediction of miRNA genes through two case studies.
SYSTEM SPECIFICATION
The ProMiR II web interface is implemented on a Linux server using PHP scripting. The core module of ProMiR, a probabilistic colearning model, is written in Java version 1.4.2. It uses the library of the program ‘RNAfold’ to predict the folding of a primary RNA sequence (Vienna RNA package version 1.6) (16). For efficient analysis and management, all data and information are stored in a MySQL database (version 5.0). The system runs on two dual 2.2 GHz OPTERON CPUs with four 1 GB RAM modules.
PRINCIPLE OF PROGRAM
ProMiR II is a web-based tool that searches for potential miRNAs in a given sequence or in its vicinity. It provides three programs: ProMiR-v, ProMiR-c and ProMiR-g. They include both common and different procedures to accomplish each purpose.
ProMiR-v searches for clusters of miRNAs near a known miRNA sequence. It maps them on one of two genome assemblies: human (hg17) or mouse (mm7) with known miRNAs and genes. ProMiR-c predicts clustered miRNAs near an miRNA candidate. It also maps predicted miRNAs on one of the two genome assemblies, as does ProMiR-v. If there are clustered miRNAs, the initial candidate is tagged as a likely ‘real’ miRNA. ProMiR-v and ProMiR-c perform predictions of human and mouse miRNAs, respectively.
ProMiR-g is a general version of ProMiR (http://bi.snu.ac.kr/ProMiR/), which searches for an miRNA in a stem–loop sequence. ProMiR-g provides the prediction of all potential miRNAs in a long sequence within various model species: Homo sapiens, Mus musculus, Rattus norvegicus, Gallus gallus, Drosophila melanogaster, Drosophila pseudoobscura, Caenorhabditis elegans and Caenorhabditis briggsae. The three programs all extract stem–loops based on the filtering parameters by scanning a given sequence with a predefined window size (range 70–150 nt) and a given shift size (range 3–10 nt). The orientation of a given sequence is determined according to the orientation of the input query (a known miRNA or a candidate sequence) in ProMiR-v and ProMiR-c. During the scanning sequence, they search for miRNA candidates beyond a set threshold of the ProMiR score, which is generated by a probabilistic model learned here with real training data based on published miRNAs (miRBase release 7.0; http://microrna.sanger.ac.uk/sequences/). In addition, ProMiR-v and ProMiR-c can find both conserved and nonconserved miRNAs across the human and mouse genome using conserved sequence information; however, ProMiR-g does not use this because it searches for unrelated miRNAs on a given sequence. For genome mapping, ProMiR-v retrieves the genome coordination information of known miRNAs from the MySQL database, but ProMiR-c takes the position of a query sequence on a genome by BLAT searching (http://genome.ucsc.edu/cgi-bin/hgBlat).
INPUT DESIGN
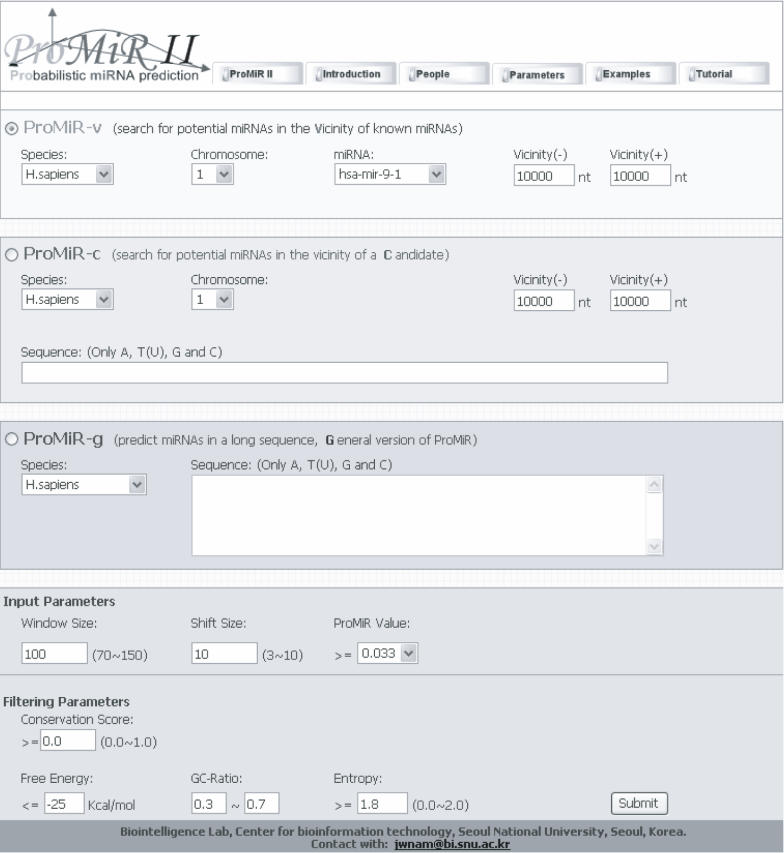
The interface of the program is shown in Figure 1. The user is required to enter different input queries according to each program. For ProMiR-v, the user selects a species (human or mouse) and one of the known miRNAs in the list box (based on miRBase release version 8.0), and enters a range to define the vicinity (up to ±10 kb). For ProMiR-c, a species is selected and a candidate sequence of 70–150 nt is input as plain text, and the range of the vicinity is then set. For ProMiR-g, a long sequence (from 70 nt to 10 kb) should be entered as plain text and one of eight species is selected as the model. In ProMiR-c and ProMiR-g, the input sequence should consist of only four bases: A, T(U), G and C. No other characters are allowed. For all programs, the user also needs to set filtering parameters and a threshold for the ProMiR score. The filtering step contains four parameters: minimum free energy (MFE), GC-ratio, entropy and conservation score (Cscore). The MFE is the cutoff value for the MFE of a stem–loop structure. The default value is −25 kcal/mol. The MFE guarantees the extraction of stem–loops with sufficient length. The G/C ratio and entropy settings filter out stem–loops made of simple repeats. The default G/C ratio ranges from 0.3 to 0.7, covering the values for most published pre-miRNAs. Entropy is entered as Shannon's entropy value, ranging from 0 to 2 (17), with a default threshold of 1.8. The Cscore uses phastCons scores for multiple alignments of eight vertebrate genomes: human (hg17), chimp (panTro1), dog (canFam1), mouse (mm5), rat (rn3), chicken (galGal2), zebrafish (danRer1) and fugu (fr1), as defined by Siepel et al. (18). The range of Cscore is from 0 to 1. If the Cscore is 0, ProMiR II will search for both conserved and nonconserved miRNAs. Otherwise, it will look for conserved miRNAs. The default Cscore is 0. ProMiR-g does not use conserved sequence information. The distribution of each parameter for published miRNAs is shown in Supplementary Figure S1.
Figure 1.
The input page of ProMiR II. Use is demonstrated in the online tutorial page (http://cbit.snu.ac.kr/~ProMiR2/tutorial.html).
ProMiR generates a score for the classification of a stem–loop. If its score is bigger than the given threshold, then ProMiR predicts that it should be an miRNA candidate. The higher the threshold the greater the specificity of classification: the lower the threshold the greater the sensitivity, as shown in the receiver operating characteristic (ROC; Supplementary Figure S2) curve. The default threshold value is 0.033.
SYSTEM OUTPUT DESIGN
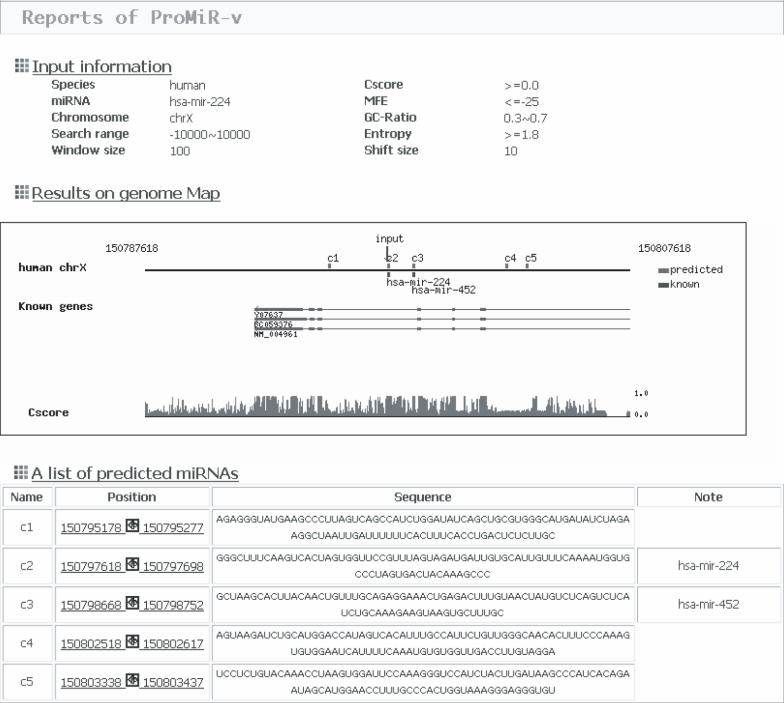
ProMiR II produces three reports (Figure 2). The first is a summary of input parameters. The next shows predicted miRNAs, known miRNAs and genes on a map. In the last, a list of miRNA candidates is displayed in order of position. The information shown for each predicted miRNA candidate includes its position, its sequence and a note. More detailed information including parameter values and a secondary structure is described in a page linked online.
Figure 2.
The output page of ProMiR II. This is also explained in the online tutorial page (http://cbit.snu.ac.kr/~ProMiR2/tutorial.html).
EXAMPLES
Clustered mouse miRNAs
To test if there are clustered miRNAs in the vicinity of a new mouse miRNA, identified by cloning and northern blotting, we applied ProMiR-c with a threshold of ProMiR score 0.017 and the default values of conservation score, entropy, MFE and G/C ratio. The search range was ±10 kb at the position of the new miRNA. The window and shift sizes were 100 and 5 nt, respectively. The program found five upstream and four known downstream clustered miRNAs, and predicted six new clustered miRNA candidates. The results are summarized in Supplementary Figure S4.
Nonrelated viral miRNAs
We analyzed a genome sequence of the human cytomegalovirus (HCMV; complete genome of strain AD169; GenBank accession no. X17403) to search for potential miRNAs using ProMiR-g. HCMV is a member of the Herpes viral family and has a double-stranded DNA genome of 229 354 bp (19). Nine miRNAs have been identified to date. Because HCMV does not have genes related to miRNA processing, it must use human genes when infecting human immune cells. Thus, because we could assume that it has the same recognition and processing mechanisms, we used the human miRNAs as training data to search for HCMV miRNAs. ProMiR-g predicted 51 candidates using a threshold ProMiR score of 0.01 and the default values of entropy, MFE and GC-ratio. The window and shift sizes were 100 and 10 nt, respectively. The candidates include five of nine published miRNAs (hcmv-mir-UL36-1, hcmv-mir-UL112-1, hcmv-mir-US5-1, hcmv-mir-US5-2 and hcmv-mir-US33-1). Results are detailed in the Supplementary Data.
DISCUSSION
ProMiR is applicable to all species given sufficient training data, and searches for related and unrelated miRNAs. Evaluation of ProMiR was performed by plotting ROCs using 5-fold cross-validation according to 15 classification thresholds (Supplementary Figure S2). ProMiR showed good performance in six species, excluding the Caenorhabditis genus.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
Acknowledgments
This work was supported by the National Research Laboratory program (M10412000095-04J0000-03610) of the Korean Ministry of Science and Technology and by a Seoul Science Fellowship from Seoul City. Funding to pay the Open Access publication charges for this article was provided by the Korean Ministry of Science and Technology.
Conflict of interest statement. None declared.
REFERENCES
- 1.Bartel D.P. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 2.Kim V.N. Small RNAs: classification, biogenesis, and function. Mol. Cells. 2005;19:1–15. [PubMed] [Google Scholar]
- 3.Lagos-Quintana M., Rauhut R., Lendeckel W., Tuschl T. Identification of novel genes coding for small expressed RNAs. Science. 2001;294:853–858. doi: 10.1126/science.1064921. [DOI] [PubMed] [Google Scholar]
- 4.Lau N.C., Lim L.P., Weinstein E.G., Bartel D.P. An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science. 2001;294:858–862. doi: 10.1126/science.1065062. [DOI] [PubMed] [Google Scholar]
- 5.Lee R.C., Ambros V. An extensive class of small RNAs in Caenorhabditis elegans. Science. 2001;294:862–864. doi: 10.1126/science.1065329. [DOI] [PubMed] [Google Scholar]
- 6.Griffiths-Jones S. The microRNA registry. Nucleic Acids Res. 2004;32:D109–D111. doi: 10.1093/nar/gkh023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ambros V., Lee R.C. Identification of microRNAs and other tiny noncoding RNAs by cDNA cloning. Methods Mol. Biol. 2004;265:131–158. doi: 10.1385/1-59259-775-0:131. [DOI] [PubMed] [Google Scholar]
- 8.Chen P.Y., Manninga H., Slanchev K., Chien M., Russo J.J., Ju J., Sheridan R., John B., Marks D.S., Gaidatzis D., et al. The developmental miRNA profiles of zebrafish as determined by small RNA cloning. Genes Dev. 2005;19:1288–1293. doi: 10.1101/gad.1310605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lai E.C., Tomancak P., Williams R.W., Rubin G.M. Computational identification of Drosophila microRNA genes. Genome Biol. 2003;4:R42. doi: 10.1186/gb-2003-4-7-r42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lim L.P., Lau N.C., Weinstein E.G., Abdelhakim A., Yekta S., Rhoades M.W., Burge C.B., Bartel D.P. The microRNAs of Caenorhabditis elegans. Genes Dev. 2003;2:2. doi: 10.1101/gad.1074403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Legendre M., Lambert A., Gautheret D. Profile-based detection of microRNA precursors in animal genomes. Bioinformatics. 2005;21:841–845. doi: 10.1093/bioinformatics/bti073. [DOI] [PubMed] [Google Scholar]
- 12.Bentwich I., Avniel A., Karov Y., Aharonov R., Gilad S., Barad O., Barzilai A., Einat P., Einav U., Meiri E., et al. Identification of hundreds of conserved and nonconserved human microRNAs. Nature Genet. 2005;37:766–770. doi: 10.1038/ng1590. [DOI] [PubMed] [Google Scholar]
- 13.Berezikov E., Guryev V., van de Belt J., Wienholds E., Plasterk R.H., Cuppen E. Phylogenetic shadowing and computational identification of human microRNA genes. Cell. 2005;120:21–24. doi: 10.1016/j.cell.2004.12.031. [DOI] [PubMed] [Google Scholar]
- 14.Xie X., Lu J., Kulbokas E.J., Golub T.R., Mootha V., Lindblad-Toh K., Lander E.S., Kellis M. Systematic discovery of regulatory motifs in human promoters and 3′ UTRs by comparison of several mammals. Nature. 2005;434:338–345. doi: 10.1038/nature03441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nam J.W., Shin K.R., Han J., Lee Y., Kim V.N., Zhang B.T. Human microRNA prediction through a probabilistic co-learning model of sequence and structure. Nucleic Acids Res. 2005;33:3570–3581. doi: 10.1093/nar/gki668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hofacker I.L. Vienna RNA secondary structure server. Nucleic Acids Res. 2003;31:3429–3431. doi: 10.1093/nar/gkg599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shannon C.E. A mathematical theory of communication. Bell Syst. Tech. J. 1948;27:379–423. [Google Scholar]
- 18.Siepel A., Bejerano G., Pedersen J.S., Hinrichs A.S., Hou M., Rosenbloom K., Clawson H., Spieth J., Hillier L.W., Richards S., et al. Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome Res. 2005;15:1034–1050. doi: 10.1101/gr.3715005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Landolfo S., Gariglio M., Gribaudo G., Lembo D. The human cytomegalovirus. Pharmacol Ther. 2003;98:269–297. doi: 10.1016/s0163-7258(03)00034-2. [DOI] [PubMed] [Google Scholar]