Abstract
GlyNest and CASPER (www.casper.organ.su.se/casper/) are two independent services aiming to predict 1H- and 13C-NMR chemical shifts of glycans. GlyNest estimates chemical shifts of glycans based on a spherical environment encoding scheme for each atom. CASPER is an increment rule-based approach which uses chemical shifts of the free reducing monosaccharides which are altered according to attached residues of an oligo- or polysaccharide sequence. Both services, which are located on separate, distributed, servers are now available through a common interface of the GLYCOSCIENCES.de portal (www.glycosciences.de). The predictive ability of both techniques was evaluated for a test set of 155 13C and 181 1H spectra of assigned glycan structures. The standard deviations between experimental and estimated shifts (1H; 0.081/0.102; 13C 0.763/0.794; GlyNest/CASPER) are comparable for both methods and significantly better than procedures where stereochemistry is not encoded. The predictive ability of both approaches is in most cases sufficiently precise to be used for an automatic assignment of NMR-spectra. Since both procedures work efficiently and require computation times in the millisecond range on standard computers, they are well suited for the assignment of NMR spectra in high-throughput glycomics projects. The service is available at www.glycosciences.de/sweetdb/start.php?action=form_shift_estimation.
INTRODUCTION
The term ‘glycomics’ describes the scientific attempt to identify and study the biological function of all the glycan molecules—the glycome—synthesized by an organism. The aim is to create a cell-by-cell catalogue of glycosyltransferase expression and detected glycan structures (1,2). Sequences for complex carbohydrates differ significantly from the simple linear one-letter code that describes genes and proteins: the number of naturally occurring residues is much larger for glycans, each pair of monosaccharide residues can be linked in several ways, and one residue can be connected to three or four others (branching). Analysis of these carbohydrates has proved difficult in the past due to their structural complexity (3,4). In comparison with the other analytical methods often used for the identification of complex carbohydrates, NMR measurements have the advantage of enabling a complete and unambiguous assignment of all structural features of glycans—the stereochemistry of monosaccharide units, the type of linkage connecting units and even their conformational preferences—using the same experimental setup.
The data produced by NMR experiments are well suited for computational approaches for two reasons. First, each NMR resonance—the so-called chemical shift of an atom given in parts per million relative to an internal or external standard—can often be assigned unambiguously to exactly one atom in a given structure. Second, the exact value of the chemical shift depends on the chemical surroundings of the atom and is essentially influenced by the type of bonds formed with the directly adjacent atoms. The influence of remotely connected atoms decreases with their distance from the focus atom. These characteristics of NMR resonances have prompted computational techniques to be used for the automatic estimation/prediction of the NMR spectra of molecules.
Increment rule based approaches use the fact that the chemical shifts of glycosyl residues in an oligo- or polysaccharide differ from those in the free monosaccharides in a predictable manner. The glycosylation shifts are additive provided that there are no steric interactions between residues more remote in sequence.
Estimation procedures based on a spherical environment encoding use a canonical linear string describing the spherical environment for each atom which is stored together with the assigned chemical shifts in lookup tables. If this procedure is repeated for a sufficient number of assigned atoms a representative set of canonical linear strings will result, which can be used to estimate shifts of new structures.
Here, we describe a web application which integrates the increment rule based approach implemented in the CASPER (5,6) program and the spherical environment encoding approach put into practice in GlyNest as part of the GLYCOSCIENCES.de portal (4), the former SWEET-DB (7). The predictive abilities of both techniques will be discussed and compared.
MATERIALS AND METHODS
Structural encoding of Glycan structures
A slightly modified version of the so-called extended form recommended by IUPAC (8) is used for the input of glycan structures in GLYCOSCIENCES.de (4). Each symbol for a monosaccharide unit is preceded by the anomeric descriptor (α and β are replaced by a and b) and the configuration symbol (D or L). The ring size is indicated by f for furanose or p for pyranose and so on. The locants of the linkage are given in parentheses between the symbols; a line indicates a linkage between two anomeric positions.
CASPER
The program CASPER (5,6) has been developed and is hosted by Stockholm University. It is an increment rule based approach that uses chemical shifts of the free reducing monosaccharides which are altered according to attached residues in an oligo- or polysaccharide sequence. Glycosylation shifts for a linkage are obtained from the chemical shifts of a disaccharide by subtracting the chemical shifts of the corresponding monosaccharides. The corrections are obtained in a similar manner by subtracting the monosaccharide and glycosylation shifts from chemical shifts of a trisaccharide. The procedure for estimating chemical shifts will be demonstrated for β-d-Glcp in β-3-O-fucosyl-lactose
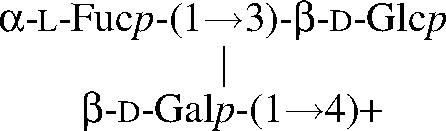 |
To calculate the chemical shifts of a glycan, one starts with the chemical shifts of the individual monosaccharides.
| C1 | C2 | C3 | C4 | C5 | C6 | |
|---|---|---|---|---|---|---|
| β-d-Glcp | 96.84 | 75.20 | 76.76 | 70.71 | 76.76 | 61.84 |
In a second step the glycosylation shifts for each linkage are added
| C1 | C2 | C3 | C4 | C5 | C6 | |
|---|---|---|---|---|---|---|
| α-l-Fucp-(1→3)-β-d-Glcp | −0.21 | 0.31 | 7.19 | −1.38 | −0.02 | 0.01 |
| β-d-Galp-(1→4)-β-d-Glcp | −0.17 | −0.22 | −1.46 | 9.19 | −1.14 | −0.56 |
| Result | 96.46 | 75.29 | 82.49 | 78.52 | 75.60 | 61.90 |
In the third step, corrections for vicinal substitution are added if available.
| C1 | C2 | C3 | C4 | C5 | C6 | |
|---|---|---|---|---|---|---|
| 0.27 | 1.21 | −4.20 | −4.60 | 0.73 | −0.37 | |
| Final chemical shifts | 96.73 | 76.50 | 78.29 | 73.92 | 76.33 | 60.92 |
GlyNest
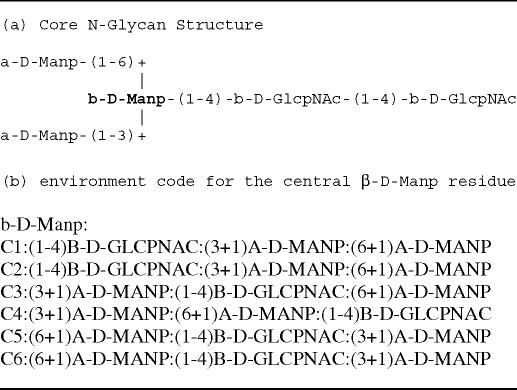
The prediction tool GlyNest has been developed and is hosted by the German Cancer Research Centre as part of the GLYCOSCIENCES.de (9) portal. It estimates chemical shifts of glycans based on a spherical environment encoding scheme. Following the general philosophy in glycobiology to describe carbohydrate structures through the topology of their monosaccharide building blocks rather than through an explicit encoding of the topology of all atoms, a residue-based spherical code was developed. It is based on a slightly modified form of the IUPAC extended form for glycan structures. Since each monosaccharide describes implicitly the stereochemistry of all stereo-centres, the resulting code reflects well the structural specialities of complex carbohydrates as required for NMR shift estimation. Table 1 demonstrates the encoding scheme for each C-atom of β-d-Manp at a branch point of the N-glycan core structure. To reflect the fact that closely connected atoms have a larger impact on the chemical shift of the atom, two rules were applied to order the list of attached residues: (i) the connected carbohydrate residues are ordered according to their increasing distance (in terms of number of bonds) from the atom to be encoded and (ii) if two distances are equal, the residue attached to the C atom with the smaller ring-atom number receives the higher priority (see Table 1 code for atom C2).
Table 1.
Stored environment code for the central β-D-Manp residue as part of the N-Glycan core region
For each atom of all structures of the GLYCOSCIENCES.de database—where assignments for the chemical shifts are available—the corresponding codes are generated and stored together with the shifts in the shift-environment table. It currently contains 27 052 1H-NMR and 14 129 13C-NMR shifts.
To perform shift estimation, the corresponding codes for each atom of the input molecule are generated and looked up in the shift-environment table. Depending on the completeness of stored codes for the unknown structure, a number of hits with differing shift values can be retrieved from the shift-environment table. The result is reported as the mean value of the stored database entries, including statistics like the standard deviation and the highest and lowest shift value found for the given code. If no match is found for a complete environment code, residues are subsequently removed from the end of the environment code and the search is repeatedly performed until hits are found. The basic requirement for the exchange of NMR data is a common chemical shift reference. GlyNest uses acetone (δH = 2.225, δC = 31.07) which is roughly equivalent to the reference used in CASPER (TSP δH = 0.00, 1,4-dioxane δC = 67.40, D2O at 70°C).
Online connection of the CASPER—GlyNest web service
CASPER and GlyNest are two services aiming to predict the chemical shifts of glycans, which are based on different scientific approaches and which are implemented on two separate hosts in Stockholm and Heidelberg. Since the World Wide Web is increasingly used for application to application communication using programmatic interfaces, it was one of the goals of this work to develop a set of XML-based descriptions to exchange the data required for NMR shift estimation. The procedure works as follows: First, GlyNest converts the IUPAC extended description to the CASPER line notation, which differ significantly in their details but not in the quality of the structural features, which are encoded. Next, GlyNest sends this description to the CASPER server using the HTTP-get mechanism. CASPER calculates the NMR spectrum, labels residues and atoms according to the nomenclature used in SugaBase (10) and returns a XML encoding of the assignments that is embedded in the HTML response. GlyNest parses the returned data and integrates the CASPER shifts into the output list.
RESULTS
The web-interface implemented in GLYCOSCIENCES.de
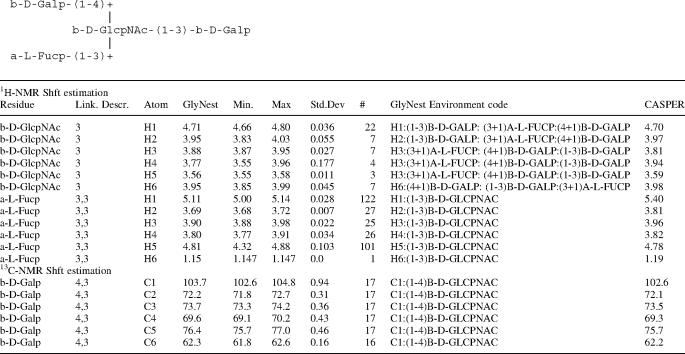
The input of glycan structures is accomplished using the extended IUPAC description in a free text editor. Table 2 shows the estimated 1H- and 13C NMR shifts for the tetrasccharide LewisX using GlyNest and CASPER. The linkage descriptor lists—starting opposite to the reducing end of the carbohydrate sequence—sequentially the number of all linkage positions (separated by commas) on the non-reducing side of each residue until the looked at residue is reached. In such a way, residues in different branches as well as at various positions within the chain can be easily and unambiguously identified. In addition the basic data used for the GlyNest estimation are displayed.
Table 2.
Example of estimated 1H- and 13C NMR-Shifts for residues of the tetrasccharide LewisX using GlyNest and CASPER
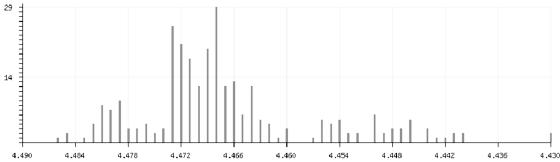
All experimental shifts found for a given environment code can be displayed in a histogram. Figure 1 shows as an example all proton shifts found in the shift-environment table for the H1 atom of terminal β-d-Galp connected 1-4 to β-d-GlcpNAc (environment code: B-D-GALP [H1:(1-4)B-D-GLCPNAC]). In addition, an option exists to display the individual structures from which a certain shift originates.
Figure 1.
Histogram of shifts stored for the environment code B-D-GALP [H1:(1-4)B-D-GLCPNAC].
Predictive Ability of GlyNest and CASPER
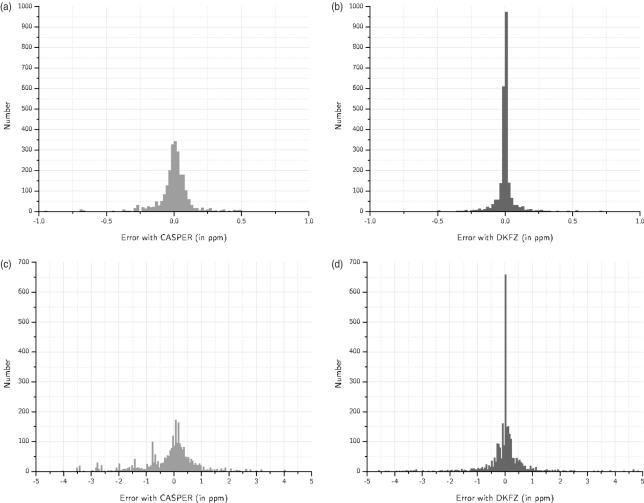
To evaluate the predictive ability of both techniques a test set of 155 13C and 181 1H spectra were selected form the GLYCOSCIENCES.de NMR database for which completely assigned glycan structures (C1 to C6, and H1 to H6) are available and which could all be estimated using GlyNest as well as CASPER. 1H spectra. N- and O- Glycans as well as glycosphingolipid head groups and polysaccharides were covered in the test set. For GlyNest estimations, the entries in the shift-environment table originating from the glycan structure to be estimated were removed. Figure 2a–d displays the respective distributions of the deviations and Table 3 reports their corresponding statistical characteristics.
Figure 2.
(a) CASPER, histogram of the deviations between estimated and experimental 1H shifts. (b) GlyNest, histogram of the deviations between estimated and experimental 1H shifts. (c) CASPER, histogram of the deviations between estimated and experimental 13C shifts. (d) GlyNest, histogram of the deviations between estimated and experimental 13C shifts.
Table 3.
Statistical characteristics for the predictive ability of CASPER and GlyNest to estimate 1H and 13C-NMR shifts of glycans
| 1H | 13C | |||
|---|---|---|---|---|
| GlyNest | CASPER | GlyNest | CASPER | |
| Total Number | 2370 | 2368 | 2679 | 2672 |
| Minimum error | 0 | 0 | 0 | 0 |
| Maximum error | 0.907 | 0.956 | 4.97 | 4.75 |
| Mean error | 0.036 | 0.075 | 0.433 | 0.7 |
| SD | 0.081 | 0.102 | 0.763 | 0.794 |
SUMMARY AND DISCUSSION
GlyNest as well as CASPER can calculate accurately 1H and 13C shifts of glycans. The standard deviations between experimental and estimated shifts are comparable for both methods. In many cases the discrepancy between calculated and experimental chemical shifts is as low as 0.05 p.p.m./resonance for 1H and 0.2 p.p.m./resonance for 13C which is comparable with the differences between measurements from different laboratories resulting from slightly dissimilar experimental conditions. Such a predictive ability may be sufficient to establish the structure of many oligo- and polysaccharides and is in many cases sufficiently accurate to be used for an automatic assignment of NMR-spectra. Since both procedures work efficiently and require computation times in the millisecond range on standard computers, they are well suited for the assignment of NMR spectra in high-throughput glycomics projects.
The online connection of two distributed services presented here, which provides access to two methodically different approaches to estimate 1H and 13C NMR shifts for glycans, demonstrates the capability of web services to make rapidly accessible distributed scientific data. The established connection will help to find shortcomings in both approaches, detect mistakes in the underlying data and thus improve the quality of both services, which will definitively lead to a better worldwide acceptance of both services within the community of glycoscienists.
Acknowledgments
The development of GlyNest at the German Cancer Research Centre was supported by a Research Grant of the German Research Foundation (DFG BIB 46 HDdkz 01-01) within the digital library program. Funding to pay the Open Access publication charges for this article was provided by DFG.
Conflict of interest statement. None declared.
REFERENCES
- 1.Raman R., Raguram S., Venkataraman G., Paulson J., Sasisekharan R. Glycomics: an integrated systems approach to structure-function relationships of glycans. Nat. Methods. 2005;2:817–824. doi: 10.1038/nmeth807. [DOI] [PubMed] [Google Scholar]
- 2.Lowe J., Marth J. A genetic approach to Mammalian glycan function. Annu. Rev. Biochem. 2003;72:643–991. doi: 10.1146/annurev.biochem.72.121801.161809. [DOI] [PubMed] [Google Scholar]
- 3.von der Lieth C.W., Bohne-Lang A., Lohmann K.K., Frank M. Bioinformatics for glycomics: status, methods, requirements and perspectives. Brief Bioinform. 2004;5:164–178. doi: 10.1093/bib/5.2.164. [DOI] [PubMed] [Google Scholar]
- 4.von der Lieth C.W., Lutteke T., Frank M. The role of informatics in glycobiology research with special emphasis on automatic interpretation of MS spectra. Biochim Biophys Acta. 2006;1760:568–577. doi: 10.1016/j.bbagen.2005.12.004. Epub 2005 Dec 29. [DOI] [PubMed] [Google Scholar]
- 5.Jansson P., Kenne L., Widmalm G. CASPER: a computer program used for structural analysis of carbohydrates. J. Chem. Inf. Comput. Sci. 1991;31:508–516. doi: 10.1021/ci00004a013. [DOI] [PubMed] [Google Scholar]
- 6.Stenutz R., Jansson P., Widmalm G. Computer-assisted structural analysis of oligo- and polysaccharides: an extension of CASPER to multibranched structures. Carbohydr Res. 1998;306:11–17. doi: 10.1016/s0008-6215(97)10047-7. [DOI] [PubMed] [Google Scholar]
- 7.Loss A., Bunsmann P., Bohne A., Loss A., Schwarzer E., Lang E., von der Lieth C.W. SWEET-DB: an attempt to create. Nucleic Acids Res. 2002;30:405–408. doi: 10.1093/nar/30.1.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.McNaught A. Nomenclature of Carbohydrates. Carbohydr. Res. 1997;297:1–90. doi: 10.1016/s0008-6215(97)83449-0. [DOI] [PubMed] [Google Scholar]
- 9.Lutteke T., Bohne-Lang A., Loss A., Goetz T., Frank M., von der Lieth C.W. GLYCOCIENCES.de: an internet portal to support glycomics and glycobiology research. Glycobiology. 2006;16:71R–81R. doi: 10.1093/glycob/cwj049. Epub 2005 Oct 20. [DOI] [PubMed] [Google Scholar]
- 10.van Kuik J.A., Hard K., Vliegenthart J.F. A 1H NMR database computer program for the analysis of the primary structure of complex carbohydrates. Carbohydr. Res. 1992;235:53–68. doi: 10.1016/0008-6215(92)80078-f. [DOI] [PubMed] [Google Scholar]