Abstract
The MPI Bioinformatics Toolkit is an interactive web service which offers access to a great variety of public and in-house bioinformatics tools. They are grouped into different sections that support sequence searches, multiple alignment, secondary and tertiary structure prediction and classification. Several public tools are offered in customized versions that extend their functionality. For example, PSI-BLAST can be run against regularly updated standard databases, customized user databases or selectable sets of genomes. Another tool, Quick2D, integrates the results of various secondary structure, transmembrane and disorder prediction programs into one view. The Toolkit provides a friendly and intuitive user interface with an online help facility. As a key feature, various tools are interconnected so that the results of one tool can be forwarded to other tools. One could run PSI-BLAST, parse out a multiple alignment of selected hits and send the results to a cluster analysis tool. The Toolkit framework and the tools developed in-house will be packaged and freely available under the GNU Lesser General Public Licence (LGPL). The Toolkit can be accessed at http://toolkit.tuebingen.mpg.de.
INTRODUCTION
As this special issue shows, the number of public bioinformatic tools and web servers is growing quickly. However, the wealth of powerful tools and servers is, in our opinion, only utilized by a fraction of biologists who would be able to profit from them. Especially for non-experts it can be very time-consuming to find out which services exist, what they can or cannot do, how to use them and how to feed results from one service to the next in the right format. This has spawned the development of two classes of servers. The first class, exemplified by PredictProtein (1), accepts a single sequence as input, runs a whole set of standard protein analysis tools and returns the bare, concatenated results in a single Email or Web page, requiring users to be familiar with the tools and their output format. The second class offers a collection of web interfaces to local versions of public bioinformatic tools. For instance, PAT (protein analysis toolkit) (2) facilitates the combination of different analysis methods by automating repetitive data processing tasks. However, its user interface and the lack of an integrated help system make PAT, suited primarily for users with biocomputing experience. Two further servers designed as toolboxes for sequence analysis are the Biology Workbench (3), which has not been updated for quite some time, and AnaBench (4), which is more geared toward analysis of DNA data.
The primary aim in developing the MPI Bioinformatics Toolkit was to offer a web service that is as easy to use as possible and that integrates a selected set of most useful methods for the analysis of protein sequences. From our own experience as users of the toolkit, its main advantages are as follows:
In-house tools: Several programs developed in our group are available only through our toolkit, e.g. HHpred (5), HHrep (6), HHsenser (7), REPPER (8), CLANS (9) and Blammer (10) (see Table 1).
Enhanced functionality of public tools: Many tools offer additional functionality compared with the original public server (see tool descriptions below).
User databases: Users may upload customized databases which are then accessible throughout the whole toolkit (upload once, use many times).
Interconnectivity: Most of the tools in the Toolkit are interconnected, allowing job results of one tool to be forwarded as input to others.
Streamlined, uniform user interface: Input forms are kept as simple and self-explanatory as possible with a uniform design and logic for all tools.
Straightforward navigation: Tools are grouped into color-coded sections that are easily accessible via tabs.
Job management: A dedicated jobs sidebar provides information and quick access to all job results of the current session.
Personal work space: Users may register and log in to gain access to a personal work space featuring long-term storage of jobs.
Table 1.
Overview of tools
| Tool | Source references | Description |
|---|---|---|
| Search | ||
| NucleotideBLAST† | Altschul et al. (11) | Sequence search against nucleotide databases (blastn, tblast, tblastx) |
| ProteinBLAST† | Altschul et al. (11) | Sequence search against protein databases (blastpgp1, blastx) |
| PSI-BLAST† | Altschul et al. (12) | Iterated sequence search against protein databases |
| fastHMMER† | Eddy (13) | Fast profile HMM search tool derived from HMMER |
| HHpred* | Söding et al. (5) | Sensitive protein homology detection, function and structure prediction by HMM-HMM comparison |
| HHsenser* | Söding et al. (7) | Sensitive iterative sequence search based on HMM-HMM comparison |
| PatternSearch* | Unpublished | Search for sequences containing a given pattern |
| Alignment | ||
| ClustalW | Thompson et al. (14) | Multiple alignment program for protein and DNA sequences |
| MUSCLE | Edgar (16) | Multiple alignment program for protein sequences |
| ProbCons | Do et al. (15) | Multiple alignment program for protein sequences |
| MAFFT | Katoh et al. (17) | Multiple alignment program for protein and DNA sequences |
| Blammer* | Frickey and Lupas (10) | Converts BLAST/PSI-BLAST output to a multiple alignment by realigning gapped regions with Clustal and removing local inconsistencies through comparison to a HMM |
| HHalign* | Söding (30) | Comparison of two alignments using HMMs |
| Sequence Analysis | ||
| HHrep* | Söding et al. (6) | Sensitive de novo repeat identification in protein sequences by HMM-HMM comparison |
| PCOILS* | Lupas et al.(31) | Coiled-coil prediction |
| REPPER* | Gruber et al. (8) | Identification of repeats and their periodicity by Fourier transform and internal sequence comparisons |
| TPRpred* | Unpublished | Prediction of TPRs (Tetratrico Peptide Repeats) and related repeats (Pentatrico Peptide Repeats and SEL1-like) |
| Aln2Plot* | Unpublished | Graphical overview of average hydrophobicity and side chain volume in a multiple alignment |
| Secondary Structure | ||
| Quick2D* | Unpublished | Concise overview of secondary structure prediction by PSIPRED (18), JNET (19) and PROFKing (20); of coiled-coils by COILS (31); of transmembrane helices by MEMSAT2 (21) and HMMTOP (22) and of natively disordered regions by DISOPRED2 (23) |
| Alignment Viewer* | Unpublished | Annotate an alignment with individual PSIPRED (18) and MEMSAT2 (21) predictions |
| Tertiary Structure | ||
| Modeller† | Sali et al. (24) | Comparative protein structure modeling by satisfying of spatial restraints |
| HHpred* | Söding et al. (5) | Sensitive protein homology detection, function and structure prediction by HMM-HMM comparison |
| Classification | ||
| PHYLIP-NEIGHBOR | Felsenstein (27) | Modules of the phylogenetic analysis package Phylip which allow the construction of distance-based, neighbor-joining trees |
| CLANS* | Frickey and Lupas (9) | Clustering tool based on all-against-all BLAST comparisons |
| ANCESCON | Cai et al. (28) | Distance-based phylogenetic inference and reconstruction of ancestral protein sequences |
| Utilities | ||
| Reformat* | Unpublished | Sequence reformatting utility |
| 6FrameTranslation* | Unpublished | Six-frame translation of nucleotide sequences |
| Extract_gis* | Unpublished | Extraction of gi-numbers from BLAST files |
| RetrieveSeq* | Unpublished | Sequence retrieval from the nr or nt database using a list of identifiers |
| gi2Promotor* | Unpublished | Extraction of nucleotide sequences upstream of genes identified by the gi-numbers of their encoded proteins |
| Backtranslator* | Unpublished | Reverse translation of amino acids into nucleotide sequences |
An asterisk after the toolname indicates that the tool was developed in our group.
A dagger indicates a public tool with extended functionality.
WEB INTERFACE
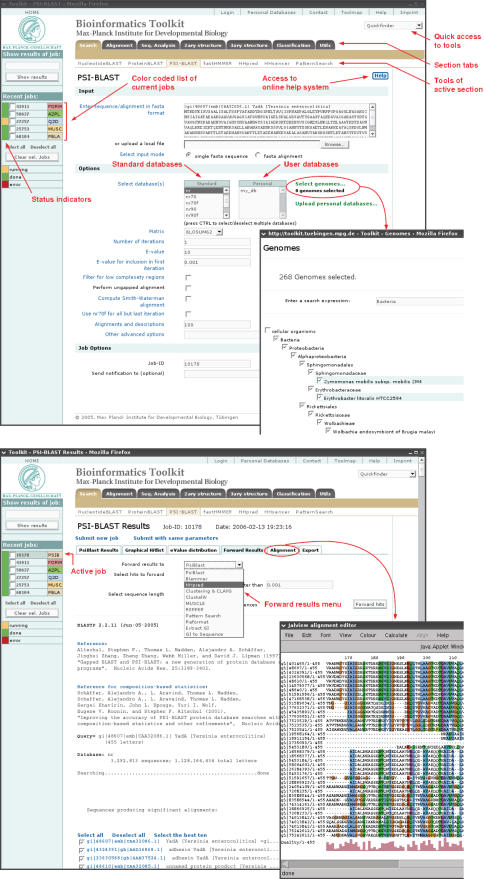
Currently, 30 bioinformatics tools and utilities can be launched from the MPI Bioinformatics Toolkit (Table 1). All tool sections are accessible from a tabbed menu bar located at the top of the page (Figure 1). Each tab reveals a submenu containing the section-specific tools, an overview page with brief descriptions for each tool and a list of selected links. Located on the left of the screen is a sidebar pane that holds a status and section-coded list of all recent jobs in the current session. One can click on previously submitted jobs to check their status and view their results. Users can also choose their own job names to organize their work. Each tool has a separate input page with a web form, in which the user can input sequence data, upload sequence files, and specify options.
Figure 1.
Input and result pages of PSI-BLAST with overlaid windows for genome databases and Jalview alignment viewer (32).
TOOL SECTIONS
The search section contains popular search tools, such as NucleotideBLAST, ProteinBLAST (11), PSI-BLAST (12), and HMMER (13), as well as our in-house developments such as HHpred, HHsenser and PatternSearch. In comparison with the NCBI server, our BLAST tools offer greater flexibility and functionality: searches can be run against uploaded personal databases or selectable sets of genomes (updated weekly from NCBI and ENSEMBL), databases can be switched between PSI-BLAST runs, alignments can be extracted, viewed online or forwarded to other tools, and two graphs show matched regions and E-value distributions. The fastHMMER tool performs HMMER searches of all standard sequence databases in ∼10% of the time by reducing the database with one iteration of PSI-BLAST at a cut-off E-value of 10 000. PatternSearch identifies sequences containing a user-defined Prosite pattern or regular expression. HHpred is a new server for protein structure and function prediction (5). It takes a query sequence as input and searches user-selected databases for homologs with a new and very sensitive method based on pairwise comparison of hidden Markov models (HMMs). Available databases, among others, are InterPro, CDD and an aligment database we build from Protein Data Bank (PDB) sequences and which can be used for 3D structure prediction. HHsenser is a transitive search method based on HMM-HMM comparison (7). This method utilizes a sequence as input and builds an alignment with as many near or remote homologs as possible, often covering the whole protein superfamily.
The alignment section includes the well-known, popular multiple alignment program ClustalW (14), together with the more recently developed multiple alignment methods ProbCons (15), MUSCLE (16) and MAFFT (17). Also in this section is Blammer (10), which converts BLAST or PSI-BLAST output to a multiple alignment by realigning gapped regions using ClustalW and removing local inconsistencies through comparison with an HMM. HHalign aligns two alignments with each other by pairwise comparison of HMMs and displays similarities in a profile–profile dotplot.
In the sequence analysis section, we have grouped tools for repeat identification and analysis of periodic regions in proteins. HHrep is a server for de novo repeat detection that is very sensitive in finding proteins with strongly diverged repeats, such as TIM barrels and β-propellers (6). REPPER (8) analyzes regions with short gapless repeats in protein sequences. It finds periodicities by Fourier transform and internal sequence similarity. The output is complemented by coiled-coil prediction and secondary structure prediction using PSIPRED (18). Aln2Plot shows a graphical overview of average hydrophobicity and side chain volume in a multiple alignment.
In the secondary structure section, Quick2D integrates the results of various secondary structure prediction programs, such as PSIPRED (18), JNET (19) and PROFKing (20), the transmembrane prediction of MEMSAT2 (21) and HMMTOP (22) and the disorder prediction of DISOPRED (23) into a single colored view. The AlignmentViewer clusters sequences by a sequence idenity criterion, annotates groups of sequences using PSIPRED and MEMSAT2 predictions of a multiple alignment and graphically displays the results in an interactive Java applet.
The tertiary structure section contains Modeller (24) and HHpred (5). Modeller is a very popular program for comparative modeling. It generates a 3D structural model from a sequence alignment of a protein sequence with one or more structural templates. In contrast to the standalone version of Modeller, the input format does not need to be PIR but can also be FASTA or most other standard multiple alignment formats. Modeller is tightly integrated with HHpred, allowing selected hits of HHpred results to be used as templates for subsequent comparative modeling. On the results page, models can be evaluated by using a browser-embedded 3D-viewer and charts with output from several model quality assessment programs are provided. This allows fast interactive refinement cycles of the underlying multiple sequence alignment. The page also provides a link to the iMolTalk server, which offers several additional tools for the detailed analysis of structures and models (25,26).
In the classification section, we offer modules of the widely used phylogenetic analysis suite PHYLIP (27), the ANCESCON package (28) for distance bases phylogenetic analysis and CLANS (9). CLANS clusters user-provided sequences based on BLAST pairwise similarities (29). The results can be analysed with a CLANS Java applet or can br exported to CLANS format.
Finally, in the utilities section there is a collection of tools which help to perform simple tasks that the user will often be confronted with. It includes a sequence reformatting utility, a six-frame translation tool for nucleotide sequences, Extract_gis for the extraction of gi-numbers from BLAST files, the RetrieveSeq tool for identifier-based sequence retrieval from the non-redundant protein or nucleotide databases at NCBI, gi2Promotor for the extraction of nucleotide sequences upstream of genes identified by the gi-numbers of their encoded proteins and a backtranslation tool.
FUTURE PLANS
Our own research on protein evolution now heavily depends on the toolkit server. We will therefore continue to integrate new tools as they become available and improve the usability of the toolkit. For instance, a project manager will be added that will further facilitate the organization and long-term storage of job results. On the technical side, we are currently in the process of porting the Toolkit to a new Rails-based web framework that permits shorter development cycles and more flexible tool interactions. The new architecture is fully object oriented and renders the Toolkit easily installable. We will package the Toolkit framework together with our in-house tools and distribute it freely under the GNU LGPL.
Acknowledgments
We thank Pawel Szczesny for contributing Aln2Plot and Tancrd Frickey for many fruitful discussions and developing various tools. We thank all users who helped to improve our server with their questions, feedback, bug reports and tool suggestions. Funding to pay the Open Access publication charges for this article was provided by the Max-Planck society.
Conflict of interest statement. None declared.
REFERENCES
- 1.Rost B., Liu J. The PredictProtein server. Nucleic Acids Res. 2003;31:3300–3304. doi: 10.1093/nar/gkg508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gracy J., Chiche L. PAT: a protein analysis toolkit for integrated biocomputing on the web. Nucleic Acids Res. 2005;33(Suppl. 2):W65–W71. doi: 10.1093/nar/gki455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Subramaniam S. The biology workbench—a seamless database and analysis environment for the biologist. Proteins. 1998;32:1–2. [PubMed] [Google Scholar]
- 4.Badidi E., De Sousa C., Lang B., Burger G. Anabench: a web/corba-based workbench for biomolecular sequence analysis. BMC Bioinformatics. 2003;4:63. doi: 10.1186/1471-2105-4-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Söding J., Biegert A., Lupas A.N. The HHpred interactive server for protein homology detection and structure prediction. Nucleic Acids Res. 2005;33:W244–W248. doi: 10.1093/nar/gki408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Söding J., Remmert M., Biegert A. HHrep: de novo protein repeat detection and the origin of TIM barrels. Nucleic Acids Res. 2006 doi: 10.1093/nar/gkl130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Söding J., Biegert A., Remmert M., Lupas A. HHsenser: exhaustive transitive profile search using HMM-HMM comparison. Nucleic Acids Res. 2006 doi: 10.1093/nar/gkl195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gruber M., Söding J., Lupas A. REPPER—repeats and their periodicities in fibrous proteins. Nucleic Acids Res. 2005;33:W239–W243. doi: 10.1093/nar/gki405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Frickey T., Lupas A. CLANS: a Java application for visualizing protein families based on pairwise similarity. Bioinformatics. 2004;20:3702–3704. doi: 10.1093/bioinformatics/bth444. [DOI] [PubMed] [Google Scholar]
- 10.Frickey T., Lupas A.N. PhyloGenie: automated phylome generation and analysis. Nucleic Acids Res. 2004;32:5231–5238. doi: 10.1093/nar/gkh867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Altschul S., Gish W., Miller W., Meyers E., Lipman D. Basic Local Alignment Search Tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 12.Altschul S., Madden T., Schaffer A., Zhang J., Zhang Z., Miller W., Lipman D. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Eddy S. Profile hidden Markov models. Bioinformatics. 1998;14:755–763. doi: 10.1093/bioinformatics/14.9.755. [DOI] [PubMed] [Google Scholar]
- 14.Thompson J., Higgin D., Gibson T. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucl. Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Do C.B., Mahabhashyam M.S., Brudno M., Batzoglou S. ProbCons: probabilistic consistency-based multiple sequence alignment. Genome Res. 2005;15:330–340. doi: 10.1101/gr.2821705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Edgar R.C. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucl. Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Katoh K., Misawa K., Kuma K.-I., Miyata T. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002;30:3059–3066. doi: 10.1093/nar/gkf436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jones D. Protein secondary structure prediction based on position-specific scoring matrices. J. Mol. Biol. 1999;292:195–202. doi: 10.1006/jmbi.1999.3091. [DOI] [PubMed] [Google Scholar]
- 19.Cuff J., Barton G. Application of multiple sequence alignment profiles to improve protein secondary structure prediction. Proteins. 2000;40:502–511. doi: 10.1002/1097-0134(20000815)40:3<502::aid-prot170>3.0.co;2-q. [DOI] [PubMed] [Google Scholar]
- 20.Ouali M., King R. Cascaded multiple classifiers for secondary structure prediction [In Process Citation] Protein Sci. 2000;9:1162–1176. doi: 10.1110/ps.9.6.1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jones T., Taylor W., Thornton J. A model recognition approach to the prediction of all-helical membrane protein structure and topology. Biochemistry. 1994;33:3038–3049. doi: 10.1021/bi00176a037. [DOI] [PubMed] [Google Scholar]
- 22.Tusnády G., Simon I. Principles governing amino acid composition of integral membrane proteins: application to topology prediction. J. Mol. Biol. 1998;283:489–506. doi: 10.1006/jmbi.1998.2107. [DOI] [PubMed] [Google Scholar]
- 23.Ward J., Sodhi J., McGuffin L., Buxton B., Jones D. Prediction and functional analysis of native disorder in proteins from the three kingdoms of life. J. Mol. Biol. 2004;337:635–645. doi: 10.1016/j.jmb.2004.02.002. [DOI] [PubMed] [Google Scholar]
- 24.Sali A., Potterton L., Yuan F., vanVlijmen H., Karplus M. Evaluation of comparative protein modeling by MODELLER. Proteins. 1995;23:318–326. doi: 10.1002/prot.340230306. [DOI] [PubMed] [Google Scholar]
- 25.Diemand A.V., Scheib H. MolTalk—a programming library for protein structures and structure analysis. BMC Bioinformatics. 2004;5:39. doi: 10.1186/1471-2105-5-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Diemand A.V., Scheib H. iMolTalk: an interactive, internet-based protein structure analysis server. Nucleic Acids Res. 2004;32:W512–W516. doi: 10.1093/nar/gkh403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Felsenstein J. PHYLIP—Phylogeny Inference Package (Version 3.2) Cladistics. 1989;5:164–166. [Google Scholar]
- 28.Cai W., Pei J., Grishin N.V. Reconstruction of ancestral protein sequences and its applications. BMC Evol. Biol. 2004;4:33. doi: 10.1186/1471-2148-4-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Frickey T., Lupas A.N. Phylogenetic analysis of AAA proteins. J. Struct. Biol. 2004;146:2–10. doi: 10.1016/j.jsb.2003.11.020. [DOI] [PubMed] [Google Scholar]
- 30.Söding J. Protein homology detection by HMM-HMM comparison. Bioinformatics. 2005;21:951–960. doi: 10.1093/bioinformatics/bti125. [DOI] [PubMed] [Google Scholar]
- 31.Lupas A., Van Dyke M., Stock J. Predicting Coiled Coils from Protein Sequences. Science. 1991;252:1162–1164. doi: 10.1126/science.252.5009.1162. [DOI] [PubMed] [Google Scholar]
- 32.Clamp M., Cuff J., Searle S.M., Barton G.J. The Jalview Java alignment editor. Bioinformatics. 2004;20:426–427. doi: 10.1093/bioinformatics/btg430. [DOI] [PubMed] [Google Scholar]