Abstract
FootPrinter3 is a web server for predicting transcription factor binding sites by using phylogenetic footprinting. Until now, phylogenetic footprinting approaches have been based either on multiple alignment analysis (e.g. PhyloVista, PhastCons), or on motif-discovery algorithms (e.g. FootPrinter2). FootPrinter3 integrates these two approaches, making use of local multiple sequence alignment blocks when those are available and reliable, but also allowing finding motifs in unalignable regions. The result is a set of predictions that joins the advantages of alignment-based methods (good specificity) to those of motif-based methods (good sensitivity, even in the presence of highly diverged species). FootPrinter3 is thus a tool of choice to exploit the wealth of vertebrate genomes being sequenced, as it allows taking full advantage of the sequences of highly diverged species (e.g. chicken, zebrafish), as well as those of more closely related species (e.g. mammals). The FootPrinter3 web server is available at: http://www.mcb.mcgill.ca/~blanchem/FootPrinter3.
INTRODUCTION
Phylogenetic footprinting is a comparative genomics approach to the computational prediction of transcription factor binding sites [for a review see (1)]. Under the premise that functional regions of a DNA sequence tend to evolve at a lower rate than non-functional regions, highly conserved regions found in a set of homologous promoter sequences are likely to be regulatory elements.
While several phylogenetic footprinting methods have been developed for identifying putative binding sites for transcription factors with known position weight matrices [e.g. rVISTA (2) and ConSite (3)], our focus here is on de novo methods, where no prior knowledge about the transcription factors involved is assumed. De novo phylogenetic footprinting methods can be separated into two groups. The first group, called alignment-based methods, contains the most published variations [see, among others, PhyloVISTA (4) and PhastCons (5)]. These methods start by computing a multiple sequence alignment of the set of orthologous sequences considered. The alignment is then scanned to identify conserved regions. Methods from the second group, called motif-based methods, do not assume that the orthologous sequences can be reliably aligned, but instead directly attempt to identify sets of subsequences that exhibit a high degree of conservation [see, e.g. FootPrinter2 (6,7)]. Here, we introduce FootPrinter3, a new web-based program that unifies the two approaches, taking advantage of the strengths of both methods.
Alignment-based versus motif-based phylogenetic footprinting
For both types of methods to clearly differentiate between binding sites and their non-functional surroundings, the set of sequences to be considered should cover a sufficiently large total amount of divergence for non-functional regions to have accumulated significantly more mutations than functional regions. This can be achieved either through a large set of relatively closely related species, or through a smaller set of more highly diverged species. However, highly diverged sequences are difficult to align. In fact, even if the set of orthologous sequences considered contains a few short, highly conserved substrings, alignment programs will often fail to align them correctly, because of the high noise of the surrounding poorly conserved sequences. In practice, this means, for example, that to identify transcription factor binding sites in a human sequence, one may want to compare it to other mammalian sequences, but comparing it to other more distantly related vertebrate species is likely to fail because of incorrect (or unavailable) alignment. In particular, the evolutionary distance between the mammalian genomes and the recently sequenced chicken genome is too large to be able to reliably align most non-coding regions (8).
Motif-based approaches have been developed precisely to address this problem, and they do not suffer from the presence of highly diverged sequences, as long as these sequences still contain the short conserved regions likely to correspond to binding sites. The main drawback of these approaches is a loss of specificity. This is due to the fact that since each motif is identified independently and outside of its context, it is possible that a set of conserved substrings identified may not be orthologous (i.e. they are not derived from a common ancestor), and that their apparent similarity is simply due to chance.
A unified approach
In this paper, we introduce FootPrinter3, a web server for the detection of short, conserved regions likely to be transcription factor binding sites in a set of partially aligned sequences. Partially aligned sequences consist of a set of ordered local alignments, each involving regions from a subset (or all) of input sequences. Not all nucleotides of all sequences need to belong to an alignment block, and regions that cannot be reliably aligned are simply left unaligned. These alignment blocks are then used as a guide for the detection of conserved substrings using a modification of the FootPrinter motif-finding algorithm (7). The motifs reported can be located both in aligned and unaligned regions and their position is guaranteed to be compatible (in a sense to be defined below) with the set of local alignment blocks. This results in a motif prediction algorithm that retains the strengths of both approaches while correcting for their weaknesses. In completely alignable sequences, our approach is similar to existing sliding-window approaches, while in completely unalignable sequences, it is equivalent to the FootPrinter2 motif-discovery algorithm (7). In between, it uses the local alignments to reduce the likelihood of finding sets of substrings that are not true orthologs, thereby increasing the specificity of the predictions.
THE FOOTPRINTER3 WEB SERVER
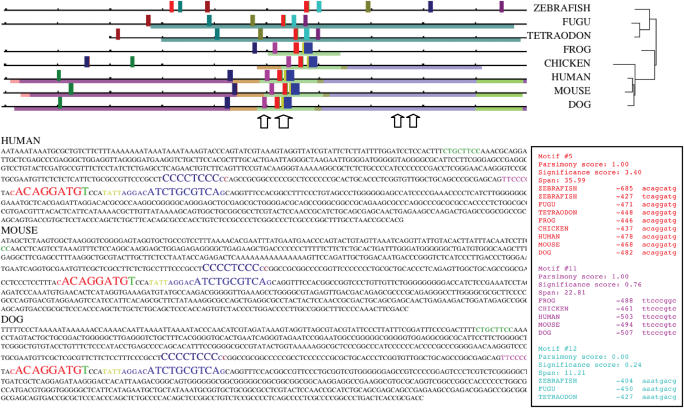
The FootPrinter3 web server allows researchers to identify, in a set of orthologous genomic sequences, short conserved regions that are likely to be regulatory elements. The user provides as input a set of unaligned orthologous nucleotide sequences, in fasta format. Typical inputs would contain from 3 to 20 orthologous (or paralogous) promoter regions of ∼1 kb each. For example, the results in Figure 1 were obtained by using the 1 kb promoter regions of the FOS gene, an important leucine-zipper transcription factor involved in apoptosis, in human, mouse, dog, chicken, frog, Tetraodon, Fugu and zebrafish. Each sequence should be labeled with the name of the species it comes from. This allows the server to automatically establish the phylogenetic relationships between the input sequences, using previously published phylogenetic trees (e.g. Tree of Life Project). In cases where the sequences come from organisms absent from our phylogenetic database, or if the sequences contain paralogs, the user is asked to provide a phylogenetic tree in Newick format.
Figure 1.
Output of the FootPrinter3 web server when run on a set of vertebrate promoter regions of the FOS gene. Each orthologous sequence extends 1 kb upstream of the start codon (in most species, the 5′-untranslated region 5′-UTR extends over ∼150 bp). Full length sequence for chicken was not available. Alignment blocks are shown in thin colored lines, while the motifs found appear as colored bars. Arrows indicate the position of experimentally verified transcription factor binding sites from Transfac. Below the graphical output are part of the set of sequences used as input, with the motifs highlighted in the same color as in the graphical display. The list of all motifs found, with the position of each of their substrings and the score statistics are shown on the right, again with a consistent color scheme. The parameters used for the run were: motif size: 8 bp; Number of mutations allowed: 2; Maximum number of mutations per branch: 1; Use TBA alignment: ‘Yes’; Allow regulatory element losses: ‘Yes’; Spanned tree significance level: ‘Significant’; Motif loss cost: 1; The running time of the query was less than 2 min. The set of input sequences and the output files are available as example from the web server.
The first step performed by FootPrinter3 is to use the TBA multiple alignment program (9) to identify local multiple alignment blocks. An alignment block consists of two or more regions of the input sequences that share significant sequence similarity, typically over at least 50 nt. Alignment blocks can be considered as regions that are very likely to be orthologous. In the graphical output produced by FootPrinter3, alignment blocks are represented by thin colored lines. Regions from different species belonging to the same alignment block are shown with the same color. For example, in Figure 1, the light-green block aligns ∼130 bp regions from the frog, chicken, human, dog and mouse, while other blocks are common to chicken and mammals (e.g. the blue–gray block), common only to mammals (e.g. the purple block) or only to puffer fishes Tetraodon and Fugu (aqua block). Note that no alignment is found between the fish sequences and the tetrapods sequences, nor is any alignment found in zebrafish. Besides the graphical view described here, the TBA output file is also reported to the user.
The second step of FootPrinter3 is to search for conserved motifs [sets of short (6–12 bp), highly conserved regions] among the input sequences. FootPrinter evaluates motif conservation using the parsimony score, defined as the minimal number of substitutions that need to have occurred during the evolution of the motif. A motif is consistent with a set of alignment blocks if the substrings it contains could form a new alignment block that would not violate the partial order relation defined by the existing blocks [see (9)]. For example, in Figure 1, a motif that would consist of a substring from the 5′ end of the human sequence and one from the 3′ end of the frog sequence would not be consistent with the alignment blocks, because in human it would be located to the left of the light-green alignment block, while, in frog, it would be located to the right of that same block. When a motif contains a substring inside an alignment block, then only substrings aligned to it in other species are eligible, unless they come from a species that does not contribute to the alignment block. For example, the green motif is located in an alignment block within mammals but is in an unaligned region in chicken. A formal definition of consistency is rather technical and is available in (10) (http://genome.cs.mcgill.ca/FootPrinter3.0/doc/FangTechReport.pdf).
The motifs identified by FootPrinter3 are reported both graphically and in text format. In the graphical output, each motif is reported as a set of colored bars whose heights depend on the significance of the observed conservation. In Figure 1, the red motif consists of a substring of each of the input sequences (with two candidates from zebrafish). This motif is compatible with the alignment because (i) all its tetrapod instances are aligned within the light-green block, (ii) the Fugu and Tetraodon instances are aligned within the aqua block, and (iii) the set of substrings is consistent with all the other alignment blocks. Other motifs are found in only a subset of the species (e.g. cyan motif in all three fish, dark green motif in chicken and mammals, etc.). All motifs reported are consistent with the set of alignment blocks. Compared to the output obtained using the same parameters but without the alignment constraints (Supplementary Figure S1), the output of Figure 1 has much higher specificity and improved readability. In general, we observe using the alignment constraints improve the specificity by about 30–50%, without a significant reduction in sensitivity. Much larger specificity improvements are obtained when longer orthologous sequences are used.
A discussion of other options inherited from the previous versions of FootPrinter is available in (6). The most important of these is the ability of FootPrinter to find motifs that are only present in a subset of the sequences considered. In that mode, FootPrinter will report a motif if the parsimony score of the substrings chosen is significantly low, given the divergence of the species in which it occurs [see (7) for more details]. This allows the identification of binding sites that may have been lost or turned over in certain lineages. A complete online help explains each input parameter and helps the user to choose appropriate parameter values and to interpret the results.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
Supplementary Material
Acknowledgments
The authors thank François Pepin for his help setting up this web server. The authors thank Martin Tompa for his useful comments, and Webb Miller for making TBA available to our server. Funding to pay the Open Access publication charges for this article was provided by a NSERC Discovery Grant.
Conflict of interest statement. None declared.
REFERENCES
- 1.Wasserman W.W., Sandelin A. Applied bioinformatics for the identification of regulatory elements. Nature Rev. Genet. 2004;5:276–287. doi: 10.1038/nrg1315. [DOI] [PubMed] [Google Scholar]
- 2.Loots G.G., Ovcharenko I. rVISTA 2.0: evolutionary analysis of transcription factor binding sites. Nucleic Acids Res. 2004;32:W217–W221. doi: 10.1093/nar/gkh383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sandelin A., Wasserman W.W., Lenhard B. ConSite: web-based prediction of regulatory elements using cross-species comparison. Nucleic Acids Res. 2004;32:W249–W252. doi: 10.1093/nar/gkh372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shah N., Couronne O., Pennacchio L.A., Brudno M., Batzoglou S., Bethel E.W., Rubin E.M., Hamann B., Dubchak I. Phylo-VISTA: interactive visualization of multiple DNA sequence alignments. Bioinformatics. 2004;20:636–643. doi: 10.1093/bioinformatics/btg459. [DOI] [PubMed] [Google Scholar]
- 5.Siepel A., Bejerano G., Pedersen J.S., Hinrichs A.S., Hou M., Rosenbloom K., Clawson H., Spieth J., Hillier L.W., Richards S., et al. Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome Res. 2005;15:1034–1050. doi: 10.1101/gr.3715005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blanchette M., Tompa M. Footprinter: a program designed for phylogenetic footprinting. Nucleic Acids Res. 2003;31:3840–3842. doi: 10.1093/nar/gkg606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Blanchette M., Tompa M. Discovery of regulatory elements by a computational method for phylogenetic footprinting. Genome Res. 2002;12:739–748. doi: 10.1101/gr.6902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hillier L.W., Miller W., Birney E., Warren W., Hardison R.C., Ponting C.P., Bork P., Burt D.W., Groenen K.D., Delany M.E., et al. Sequence and comparative analysis of the chicken genome provide unique perspectives on vertebrate evolution. Nature. 2004;432:695–716. doi: 10.1038/nature03154. [DOI] [PubMed] [Google Scholar]
- 9.Blanchette M., Kent W.J., Riemer C., Elnitski L., Smit A.F.A., Roskin K.M., Baertsch R., Rosenbloom K., Clawson H., Green E.D., et al. Aligning multiple genomic sequences with the threaded blockset aligner. Genome Res. 2004;14:708–715. doi: 10.1101/gr.1933104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fang F., Blanchette M. Algorithms for phylogenetic footprinting in semi-alignable sequences. Internal Technical report. 2006. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.