Abstract
DiANNA is a recent state-of-the-art artificial neural network and web server, which determines the cysteine oxidation state and disulfide connectivity of a protein, given only its amino acid sequence. Version 1.0 of DiANNA uses a feed-forward neural network to determine which cysteines are involved in a disulfide bond, and employs a novel architecture neural network to predict which half-cystines are covalently bound to which other half-cystines. In version 1.1 of DiANNA, described here, we extend functionality by applying a support vector machine with spectrum kernel for the cysteine classification problem—to determine whether a cysteine is reduced (free in sulfhydryl state), half-cystine (involved in a disulfide bond) or bound to a metallic ligand. In the latter case, DiANNA predicts the ligand among iron, zinc, cadmium and carbon. Available at: http://bioinformatics.bc.edu/clotelab/DiANNA/.
INTRODUCTION
Cysteine residues play a unique role in determining protein stability and function. Cysteines may be reduced (free, where sulfur occurs in the reactive sulfhydryl form) or oxidized; the latter may be involved in a disulfide bond, i.e. a half-cystine, or instead covalently bound to a metallic ligand that is part of a prosthetic group. Experimental determination of cysteine species (free, half-cystine, ligand-bound) is non-trivial, and often only the knowledge of the three-dimensional structure indicates the species. For this reason, cysteine classification is an important bioinformatics problem that may be approached by using machine learning methods. In this paper, we apply support vector machines (SVM) to the ternary cysteine classification problem, to determine whether a given cysteine is free, a half-cystine or ligand-bound. To the best of our knowledge, the present paper describes the only existent ternary cysteine classification program.
It is reasonable to assume that each species of cysteine resides in a distinct micro-environment which influences the cysteine redox potential and its steric accessibility. This hypothesis is confirmed and exploited in several machine learning approaches for cysteine classification that, while different, share the common feature that the discrimination is based on the analysis of the cysteine sequence context, using a symmetric sequence window of length w centered about each cysteine. Particular effort has been spent on the binary classification problem to discriminate intra-chain half-cystines from free cysteines, the latter being the most represented species. For this problem, various methods have yielded steadily increasing prediction accuracies (1,2). Nevertheless, other species of cysteines exist—namely ligand-bound cysteines and half-cystines involved in inter-chain disulfide bonds. Such cysteines reside in possibly different micro-environments, hence may be discernable from other species. Only one attempt has been made to discriminate ligand-bound cysteines; specifically, Passerini and Frasconi (3) obtained prediction accuracy of ∼90% for the binary classification problem of distinguishing ligand-bound cysteines from half-cystines.
DiANNA 1.1 is the only software which performs ternary cysteine classification; all other cysteine classification web servers consider only the binary classification problem of discriminating free cysteines from intra-chain half-cystines. In this paper, we apply a SVM with (a variant of) the spectrum kernel (4) to classify cysteines into three different species: free, half-cystine or ligand-bound. For predicted ligand-bound cysteines, we further refine the classification by predicting the bound ligand to be iron, zinc, cadmium or carbon. Although we have some results concerning inter-chain disulfide bonds (data not shown), the DiANNA web server is intended only for use with single-chain proteins.
DATASET
To test and train a ternary SVM predictor for cysteine classification, it was necessary to build a dataset, in which each cysteine species is well represented. This was done as follows. From the Protein Data Bank (5), we extracted the set of single-chain proteins containing ligand-bound cysteines, and produced a non-redundant collection by using the program UniqueProt (6) with HSSP distance set to 0. This produced a list of 202 chains, denoted by UP. To enrich the small number (60) of half-cystines examples (which is probably not representative), we considered the 967 non-redundant protein chains used in (1) for training and testing a neural network to predict cysteine oxidation state prediction (dataset MA). We merged the UP and MA datasets, and re-applied UniqueProt to eliminate redundancy between the two lists. From each redundancy cluster, we selected one member containing ligand-bound cysteines, if available (if not, we selected the representative member proposed by UniqueProt). In this fashion, we obtained a dataset (denoted UPMA) of 526 chains, with adequate representation of each of the three cysteine classes. Table 1 displays the number of cysteines in each species, and Table 2 presents the number of chains containing each species. From each protein in UPMA, we extracted symmetric windows of size w centered around each cysteine. Different values of w were tested, and the best results were obtained for w = 17 [the same value led to the best performance in (3)]. The annotated UPMA list is available at URL http://bioinformatics.bc.edu/clotelab/DiANNA/UPMA_annotated.html.
Table 1.
Total number of different cysteine species in datasets considered in this paper
| Dataset | LC | IA HC | IE HC | FC | Total |
|---|---|---|---|---|---|
| UP | 624 | 60 | 2 | 546 | 1230 |
| MA | 216 | 1481 | 37 | 2412 | 4109 |
| UPMA | 624 | 608 | 24 | 1199 | 2455 |
The description of each dataset can be found in the section ‘Dataset’. Legend: IA, intra-chain disulfide bonds; IE, inter-chain disulfide bonds; HC, half-cystines; FC, free cysteines; LC, ligand-bound cysteines.
Table 2.
Breakdown of protein chains which contain at the same time half-cystines (HC), free cysteines (FC) and ligand-bound cysteines (LC), for each of the three datasets considered in this paper
| Chains | UPMA | UP | MA |
|---|---|---|---|
| Total | 526 | 202 | 967 |
| w/ HC | 140 | 19 | 291 |
| w/ FC | 363 | 139 | 716 |
| w/ LC | 189 | 202 | 52 |
| w/ both HC and FC | 28 | 9 | 65 |
| w/ both HC and LC | 17 | 19 | 1 |
| w/ both FC and LC | 128 | 139 | 26 |
| w/ HC, FC and LC | 7 | 9 | 0 |
SVM PREDICTION USING STRING KERNELS
SVMs were introduced by Vapnik within the context of a mathematically rigorous statistical learning theory—for a very clear exposition of this topic see (7). Often demonstrating better prediction accuracy than neural networks, SVMs have become increasingly popular in bioinformatics, with applications ranging from translation initiation site determination (8), remote homology detection in proteins (9), viral protease cleavage site prediction (10), fast computation of Z-scores for minimum free energy of RNA (11) and so on.
To apply SVMs to the ternary cysteine classification problem, we use the spectrum representation (4) which describes an amino acid sequence by specifying the vector of k-mers which occur; i.e. for peptide p, define Φk(p) = 〈φa(x):a ∈ Ak〉, where φa(x) is the number of occurrences of the k-mer a in p, and A is the set of 1-letter codes of amino acids. Leslie et al. use the term spectrum kernel resp. mismatch kernel in (4,13), and Busuttil et al. use the term profile-based kernel in (14). More rigorously speaking, these authors actually apply classical kernels [e.g. the linear kernel in (4,13)] for new representations of amino acid sequences—the spectrum representation, mismatch representation, profile-based spectrum representation. In this paper, we obtained the best results when k = 3, so that the amino acid sequence p in each size w window is encoded by the vector Φ3(p) of 8000 coordinates, giving the number of occurrences of each 3-mer in p. With the spectrum representation, we used the software libSVM (12) with a degree 2 polynomial kernel, such that the cost parameter C = 1—for explanation of these parameters see (12).
To train and test the SVMs we used 5-fold cross-validation, splitting positive and negative datasets into five random subsets of approximatively the same size. Using libSVM, the SVM multiclass classifier outputs, for each cysteine in the input sequence, the probability of being a free cysteine (FC), a half-cystine (HC) and ligand-bound (LC). To measure the performance of the algorithm we used the Q3 score, which is the ratio between correctly predicted examples and the total number of examples. The Q3 score is commonly used for the performance evaluation of three states (sheet, helix, coil) secondary structure predictors—e.g. see (15). Additionally, we computed the Qp score, which is the fraction of proteins for which all cysteines are correctly classified. The results (Table 3) show that the highest Q3 and Qp scores are obtained using for the spectrum representation with a degree 2 polynomial kernel (scores of 0.78 and 0.53, respectively). Although the papers (13) and (14,16) report that the mismatch and profile-based kernels outperform the spectrum kernel in protein classification experiments, we found that this is not the case for cysteine oxidation state prediction. Additional data describing the results of binary classification experiments can be found in the web supplement at the DiANNA web site.
Table 3.
Performance measure (Q3 and Qp scores) for the three-class prediction of LC, HC, FC using different kernels and input representation
| Kernel | Q3 | Qp | ||||
|---|---|---|---|---|---|---|
| SpR | MmR | PrfR | SpR | MmR | PrfR | |
| Linear | 0.75 | 0.64 | 0.63 | 0.45 | 0.43 | 0.43 |
| Polynomial (2) | 0.78 | 0.74 | 0.74 | 0.53 | 0.46 | 0.45 |
| Polynomial (3) | 0.76 | 0.72 | 0.72 | 0.5 | 0.47 | 0.47 |
| RBF | 0.75 | 0.73 | 0.72 | 0.43 | 0.43 | 0.43 |
The Q3 score is the ratio between correct prediction and total number of examples. The Qp score is the fraction of proteins for which all cysteines are correctly predicted. Q3 and Qp scores are obtained averaging the results of a 5-fold cross validation. Optimal values of the C parameter and the γ parameter for the radial basis function (RBF) kernel are estimated by a grid search. Legend: SpR—Spectrum representation; MmR—Mismatch representation; PrfR—Profile representation.
Table 4 displays the number of examples in dataset UPMA for each distinct ligand type in ligand-bound cysteines. For the cases for which we have at least 39 examples (i.e. Zn, Fe, Cd, C) we investigated whether machine learning can be used to discriminate the atomic species bound—i.e. whether sequence context of each type of ligand is significantly different. Experiments were performed where the positive set consisted of amino acid sequences symmetrically flanking those cysteines bound to a specific ligand (say iron), while the negative set consisted of sequences flanking cysteines bound to a different ligand. In the case of cadmium (Cd) and carbon (C), we randomly resampled the positive training set (which is substantially smaller than the negative training set) until the number of positive and negative examples was the same (note that the test set is unchanged). As in ternary cysteine classification, we found that the best discrimination was obtained in using the degree 2 polynomial kernel with the spectrum representation. Results are reported in Table 5 and Figure 1.
Table 4.
Total number of distinct atomic ligands found covalently bound to cysteine residues in the UPMA dataset.
| Cys-bound atom | Examples |
|---|---|
| As | 2 |
| Au | 1 |
| C | 89 |
| Cd | 39 |
| Cu | 10 |
| Fe | 185 |
| H | 1 |
| Hg | 24 |
| Mn | 1 |
| Ni | 6 |
| Pb | 1 |
| S | 27 |
| U | 2 |
| Zn | 225 |
Table 5.
Performance measures for the prediction of cysteines bound to specific ligands
| Measure | Zn | Cd | Fe | C |
|---|---|---|---|---|
| Acc | 0.93 | 0.99 | 0.91 | 0.96 |
| Sen | 0.8 | 0.97 | 0.67 | 0.74 |
| Spe | 0.99 | 1 | 0.98 | 0.99 |
| MCC | 0.84 | 0.99 | 0.74 | 0.83 |
| AUC | 0.97 | 0.97 | 0.94 | 0.94 |
Legend: Acc—accuracy; Sen—sensitivity; Spe—specificity; MCC—Matthew's correlation coefficient; AUC—area under the ROC curve.
Figure 1.
ROC curves for the prediction of cysteines covalently bound to specific ligands. [For an explanation of receiver operating characteristic (ROC) curves see (20)].
WEB SERVER
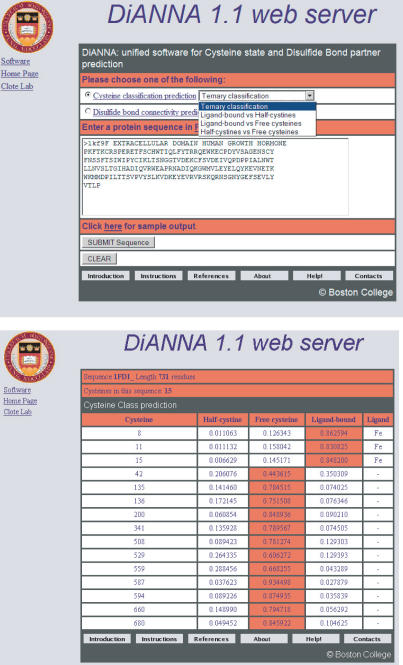
DiANNA 1.1 has a simple user-friendly web interface, which allows the user to obtain a prediction of the state (free, half-cystine or ligand-bound) for each cysteine in an input protein. The ternary SVM predictor outputs the highest probability class, and, for those cysteines predicted as ligand-bound, the most likely ligand is displayed (among iron, zinc, cadmium, carbon), by a winner-takes-all decision. Additionally, as described previously (17,18), DiANNA 1.1 uses a state-of-the-art method to predict the disulfide connectivity—i.e. which cysteines form a disulfide bond with which other cysteines. A screen shot of the DiANNA 1.1 web server output for a ternary classification prediction is shown in Figure 2. Additionally, DiANNA 1.1 allows all possible binary classification predictions for the three cysteine classes (free, half-cystine, ligand-bound). The web server interface is largely self-explanatory. The upper panel of Figure 2 displays the input form, including the pull-down menu, which allows the user to choose the classifier used for cysteine state prediction (ternary classifier, or one of three binary classifiers). The lower panel of Figure 2 displays the output of the ternary cysteine state classifier, indicating the probability of each class (half-cystine, free cysteine, ligand-bound). In the case of predicted ligand-bound cysteines, the predicted ligand is listed in the right-most column. The user enters a protein in FASTA format, possibly including a FASTA comment, and chooses either to predict the cysteine state for each cysteine, or to determine the disulfide connectivity. The latter function has already been described in (17).
Figure 2.
DiANNA ternary cysteine classification prediction input and output example. Upper panel: The DiANNA web-server update allows the user to choose between disulfide connectivity prediction and cysteine classification (ternary cysteine classification is only available in the 1.1 update). In the latter case, the user can type or paste a FASTA sequence in a text box, then choose among four different classification predictions by means of a drop down menu (i.e. the ternary LC versus HC versus FC classification, and the three binary classifications LC versus HC, LC versus FC and HC versus FC). Lower panel: Output for the ternary classification. For each cysteine in the submitted sequence, the SVM model predicts the probability of being half-cystine, free cysteine or ligand-bound. The class having the highest probability is highlighted. If a specific cysteine is predicted as ligand bound, a tentative prediction about the putative ligand (out of four possible ligands) is attempted.
CONCLUSION
Given the amino acid sequence of a protein, DiANNA (17) is a state-of-the-art method to predict disulfide connectivity topology. Version 1.0 of the DiANNA web server, described in (18), additionally predicts the oxidation state of each cysteine (free or half-cystine), by using our implementation of the neural network of Fariselli et al. (19). In version 1.1 of the DiANNA web server, described in this paper, we replace the binary classifier of (19) by a SVM with degree 2 polynomial kernel for the spectrum representation (4). Using libSVM, we obtain a ternary classifier, capable of discriminating between free cysteines, half-cystines and ligand-bound cysteines. Moreover, for the latter, DiANNA 1.1 predicts the type of ligand. To the best of our knowledge, this is the first application of string-based kernels to sequence windows; until this paper, such kernels had been used only for protein classification.
Acknowledgments
We would like to thank J. Waldispühl for helping in the web interface design, and anonymous referees for some valuable suggestions. Work of P.C. was partially supported by NSF DBI-0543506. Funding to pay the Open Access publication charges for this article was provided by NSF grant DBI-0543506.
Conflict of interest statement. None declared.
REFERENCES
- 1.Martelli P.L., Fariselli P., Malaguti L., Casadio R. Prediction of the disulfide bonding state of cysteines in proteins with hidden neural networks. Protein Eng. 2002;15:951–953. doi: 10.1093/protein/15.12.951. [DOI] [PubMed] [Google Scholar]
- 2.Chen Y.C., Lin Y.S., Lin C.J., Hwang J.K. Prediction of the bonding states of cysteines using the support vector machines based on multiple feature vectors and cysteine state sequences. Proteins. 2004;55:1036–1042. doi: 10.1002/prot.20079. [DOI] [PubMed] [Google Scholar]
- 3.Passerini A., Frasconi P. Learning to discriminate between ligand-bound and disulfide-bound cysteines. Protein Eng. Des. Sel. 2004;17:367–373. doi: 10.1093/protein/gzh042. [DOI] [PubMed] [Google Scholar]
- 4.Leslie C., Eskin E., Noble W.S. The spectrum kernel: a string kernel for SVM protein classification. Pac. Symp. Biocomput. 2002:564–575. [PubMed] [Google Scholar]
- 5.Berman H.M., Battistuz T., Bhat T.N., Bluhm W.F., Bourne P.E., Burkhardt K., Feng Z., Gilliland G.L., Iype L., Jain S., et al. The Protein Data Bank. Acta Crystallogr. D Biol. Crystallogr. 2002;58:899–907. doi: 10.1107/s0907444902003451. [DOI] [PubMed] [Google Scholar]
- 6.Mika S., Rost B. UniqueProt: creating representative protein sequence sets. Nucleic Acids Res. 2003;31:3789–3791. doi: 10.1093/nar/gkg620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vapnik V. The Nature Of Statistical Learning Theory. NY: Springer; 1995. [Google Scholar]
- 8.Zien A., Ratsch G., Mika S., Scholkopf B., Lengauer T., Muller K.R. Engineering support vector machine kernels that recognize translation initiation sites. Bioinformatics. 2000;16:799–807. doi: 10.1093/bioinformatics/16.9.799. [DOI] [PubMed] [Google Scholar]
- 9.Jaakkola T., Diekhans M., Haussler D. Using the Fisher kernel method to detect remote protein homologies. Proc. Int. Conf. Intell. Syst. Mol. Biol. 1999:149–158. [PubMed] [Google Scholar]
- 10.Narayanan A., Wu X., Yang Z.R. Mining viral protease data to extract cleavage knowledge. Bioinformatics. 2002;18:S5–S13. doi: 10.1093/bioinformatics/18.suppl_1.s5. [DOI] [PubMed] [Google Scholar]
- 11.Washietl S., Hofacker I.L., Stadler P.F. Fast and reliable prediction of noncoding RNAs. Proc. Natl Acad. Sci. USA. 2005;102:2454–2459. doi: 10.1073/pnas.0409169102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fan R.-E., Chen P.-H., Lin C.-J. Working set selection using the second order information for training SVM. J. Machine Learning Res. 2005;6:1889–1918. [Google Scholar]
- 13.Leslie C.S., Eskin E., Cohen A., Weston J., Noble W.S. Mismatch string kernels for discriminative protein classification. Bioinformatics. 2004;20:467–476. doi: 10.1093/bioinformatics/btg431. [DOI] [PubMed] [Google Scholar]
- 14.Busuttil S., Abela J., Pace G. Support vector machines with profile-based kernels for discriminative protein classification. Genome Inform. 2004;15:191–200. [PubMed] [Google Scholar]
- 15.Jones D.T. Protein secondary structure prediction based on position-specific scoring matrices. J. Mol. Biol. 1999;292:195–202. doi: 10.1006/jmbi.1999.3091. [DOI] [PubMed] [Google Scholar]
- 16.Kuang R., Ie E., Wang K., Siddiqi M., Freund Y., Leslie C. Profile-based string kernels for remote homology detection and motif extraction. Proc. IEEE Comput. Syst. Bioinform. Conf. 2004:152–160. doi: 10.1109/csb.2004.1332428. [DOI] [PubMed] [Google Scholar]
- 17.Ferrè F., Clote P. Disulfide connectivity prediction using secondary structure information and diresidue frequencies. Bioinformatics. 2005;21:2336–2346. doi: 10.1093/bioinformatics/bti328. [DOI] [PubMed] [Google Scholar]
- 18.Ferrè F., Clote P. DiANNA: a web server for disulfide connectivity prediction. Nucleic Acids Res. 2005;33:W230–W232. doi: 10.1093/nar/gki412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fariselli P., Riccobelli P., Casadio R. Role of evolutionary information in predicting the disulfide-bonding state of cysteine in proteins. Proteins. 1999;36:340–346. [PubMed] [Google Scholar]
- 20.Gribskov M., Robinson N. The use of receiver operating characteristic (ROC) analysis to evaluate sequence matching. Comput. Chem. 1996;20:25–34. doi: 10.1016/s0097-8485(96)80004-0. [DOI] [PubMed] [Google Scholar]