Abstract
RNA molecules fold into characteristic secondary structures for their diverse functional activities such as post-translational regulation of gene expression. Searching homologs of a pre-defined RNA structural motif, which may be a known functional element or a putative RNA structural motif, can provide useful information for deciphering RNA regulatory mechanisms. Since searching for the RNA structural homologs among the numerous RNA sequences is extremely time-consuming, this work develops a data preprocessing strategy to enhance the search efficiency and presents RNAMST, which is an efficient and flexible web server for rapidly identifying homologs of a pre-defined RNA structural motif among numerous RNA sequences. Intuitive user interface are provided on the web server to facilitate the predictive analysis. By comparing the proposed web server to other tools developed previously, RNAMST performs remarkably more efficiently and provides more effective and flexible functions. RNAMST is now available on the web at http://bioinfo.csie.ncu.edu.tw/~rnamst/.
INTRODUCTION
RNA molecules are involved in numerous biological processes, ranging from gene regulation to protein synthesis. Some regulatory mechanisms involve binding of proteins or metabolites to specific sites of RNA molecules (1–7). For instance, riboswitches are metabolite-binding domain within a specific mRNA, and can regulate both transcription and translation by binding their corresponding targets (4–7).
The function of RNA molecules frequently depends on the motifs conserved in both secondary structure and sequence. Numerous biologically relevant RNA motifs, including signal recognition particle, RNase P, cis-acting RNA regulatory elements (eleRNAs) and microRNAs, are found in the untranslated regions (UTRs) of mRNA and are involved in the regulation of gene expression. Most of these elements are conserved better in structures than in sequences (8–10). Searching these highly conserved RNA structural motifs can yield useful information for deciphering RNA regulatory mechanisms.
Several publicly available motif search tools, e.g. tRNAscan-SE (11) and Riboswitch finder (12), were developed to identify transfer RNA and riboswitch RNA against RNA sequence, respectively. In addition to these tools for achieving special types of regulatory RNAs, numerous tools such as Palingol (13), RNAMotif (14), PatSearch (15) and RNABOB (http://selab.wustl.edu/cgi-bin/selab.pl?mode=software#rnabob) were developed so that users could define RNA motifs according to their requirements. Palingol (13) uses a declarative programming language to describe nucleic acids' secondary structures and to scan sequence database. RNAMotif (14) devises the definition of RNA structural descriptor to describe an RNA secondary structure and scans sequences for structural homologs. PatSearch (15) is a website for detecting patterns and structural motifs in nucleotide sequences. RNABOB is a program searching for RNA motifs in sequence databases.
Although these tools can successfully define the RNA structural motifs and identify putative motifs among the limited range of RNA sequences, the search time can become a critical concern when searching in very large quantities of RNA sequences. In order to implement a web server for the convenient usage of biologists, more efficient tools for identify homologs of RNA structural motifs are crucial. This work develops an efficient and flexible tool, namely RNAMST, for rapidly searching for homologs of a pre-defined RNA structural motif, including hairpin, internal loop as well as multi-branch loop, among numerous RNA sequences. Additionally, intuitive web interface are provided on the web server to facilitate the predictive analysis.
METHODS
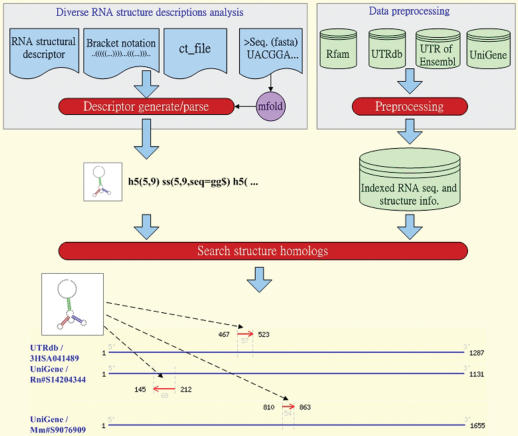
Figure 1 shows the system flow of RNAMST, which comprises RNA structural description generation, data preprocessing and searching structural homologs. Each part of the RNAMST is described in detail below.
Figure 1.
Search process of RNAMST. The figure shows how RNAMST searches a specified RNA structural motif in the given RNA sequences.
RNA structural descriptor generation
The RNAMST web server accepts four input formats, i.e. the RNA structural descriptor, bracket notation, ct format (16) and RNA sequence, to allow users to easily describe an RNA structural motif. An RNA structural descriptor defined previously by RNAMotif is used to describe a specific structural motif, and can be recognized through the step of searching structure homologs. The bracket notation and ct format are transformed to structural descriptor. In addition, the input of RNA sequence is folded by mfold (16), stored as ct format file and then transformed into structural descriptor.
Data preprocessing
The RNAMST designs a preprocessing strategy to prepare all the possible hairpins of the search sequences in advance. For each search sequence, RNAMST calculates the possible helices based on local sequence alignment. For example, Supplementary Figure S1 contains a hairpin and a non-external helix. RNAMST detects all possible hairpins and collects them into the indexed RNA sequence database. Additionally, this work exploits the multilevel index technique to establish the index file based on the order of stem length, loop length and the number of mispairs. Through effectively using these indexed files and adopting binary search algorithm, RNAMST can retrieve the requiring hairpins rapidly and efficiently.
Furthermore, since tandem repeat may produce excessive noise when determining helical regions, the tandem repeats within the search sequences are detected by the Tandem Repeat Finder (17), and then are filtered out from the indexed RNA sequence database. Following the preprocessing, RNAMST can retrieve all the requiring hairpin information in search sequences to fit the input RNA structural motifs extremely quickly.
Searching structural homologs
The RNAMST search algorithm involves two steps: the combination of external hairpins and the combination of non-external helices. Initially, RNAMST seeks all possible hairpins satisfying the constraints, which are described in RNA structural descriptor, from the indexed RNA sequence database. Subsequently, RNAMST then seeks the non-external helices using local alignment. If non-external helices are found, RNAMST integrates the external hairpins and non-external helices to fit the RNA structural motif, thus generating results of RNA structural homologs.
The RNAMST program is implemented using C++ programming language and runs under the Linux operating system on a PC server. As to the preprocessing of the search sequence database, Supplementary Table S1 gives the indexing time and required storage.
PERFORMANCE EVALUATION
This section describes the efficiency and flexibility of RNAMST. Several well-known RNA motifs in Rfam (18) are used to search for structural homologs against the sequences with different sizes, and the search time are compared with those experiments done using RNAMotif (14).
To evaluate the performance of searching for structural homologs among large numbers of sequences, RNAMotif (14) is compared with RNAMST developed here in searching for two kind of RNA motifs, iron response element (IRE) (19) and purine riboswitch (5), with different sized databases.
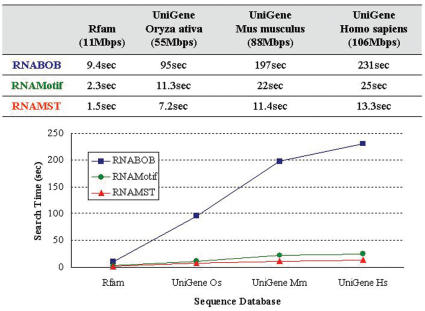
Supplementary Figure S1 shows the RNA structural motif of IRE, which is a short and conserved stem–loop found in UTRs of various mRNA. The IRE regulates mRNA translation by interacting with iron regulatory proteins (19). The RNA structural descriptor of IRE is given in Supplementary Figure S1. Figure 2 gives the comparison of the search time of RNABOB, RNAMotif and RNAMST with different sized databases. For example, RNABOB and RNAMotif spent 231 and 25 s in detecting homologs of IRE motifs in UniGene human sequence database, respectively, while our RNAMST only needed 13.3 s to accomplish the same search. Since IRE is a very small motif and is highly conserved, the constraint of the IRE structural descriptor is quite strict, making it easy to be detected. Despite this, RNAMST is still ∼9 and 1.9 times faster than RNABOB and RNAMotif, respectively, and the difference increases as the structure becomes more complex and the constraints loosen.
Figure 2.
Performance comparison for RNABOB, RNAMotif and RNAMST. Search time of RNABOB, RNAMotif and RNAMST against the different search sequence databases with the IRE motif.
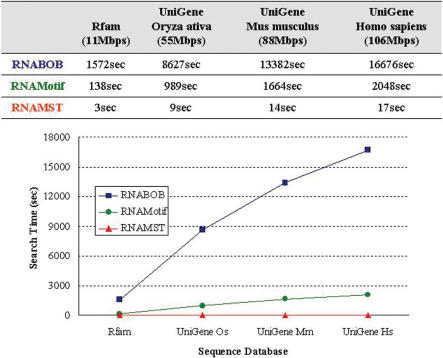
Supplementary Figure S2 gives the RNA structural descriptor of purine riboswitch, which is a metabolite-binding domain within certain mRNAs and is involved in modulating gene expression (5). Similarly, Figure 3 illustrates the comparison of the search time among RNABOB, RNAMotif and RNAMST for different sized databases. For instance, RNABOB and RNAMotif spend 16 676 and 2048 s in detecting homologs of purine riboswitch, respectively, while our RNAMST spent only 17 s. In this case, owing to the more complex structure and looser constraints of purine riboswitch compared to IRE motif, RNAMST performs ∼980 and 120 times faster than RNAMotif and RNABOB, respectively. After the above overall evaluation, our RNAMST has a lot of improvement in search performance.
Figure 3.
Performance comparison for RNABOB, RNAMotif and RNAMST. Search time of RNABOB, RNAMotif and RNAMST for the different search sequence databases with purine riboswitch motifs.
Flexible structural search
Besides the efficient RNA structural homolog search, RNAMST also consider flexible structures with asymmetric mispairs and bulges to increase its applicability and practicality. For example, to search for homologs allowing asymmetric mispairs and bulges of a long hairpin such as the example of RNA structural motif given in Supplementary Figure S3-A, RNAMotif only yields results in structures with symmetrical mispair (Supplementary Figure S3-B) but fails to find any results in cases involving asymmetrical mispair (Supplementary Figure S3-D) and bulge (Supplementary Figure S3-C, E), despite two types of structures are similar. Because searching for motifs with asymmetrical mispair or bulge is extremely time-consuming, none of the RNA structural motif search tools developed previously support this consideration, despite these structures commonly appearing in RNA secondary structures. RNAMST overcomes this problem through preprocessing so as to support more flexible search for RNA structural homologs.
This work presents an example of finding microRNA (miRNA) mir-1 motif using flexible search. The miRNAs take the form of a long hairpin structure (Supplementary Figure S4) and trigger a cis-acting translational repressive activity by involving Dicer enzyme. The RNA structural descriptor of the mir-1 is given in Supplementary Figure S4. Supplementary Figure S5 lists the sequences in mir-1 family in Rfam that can be identified using mir-1 motif by RNAMST. All seven known sequences in the mir-1 family of Rfam were identified correctly by RNAMST with flexible structure search, but only one of seven sequences, AC002442.1, was identified by RNAMotif.
INTERFACE
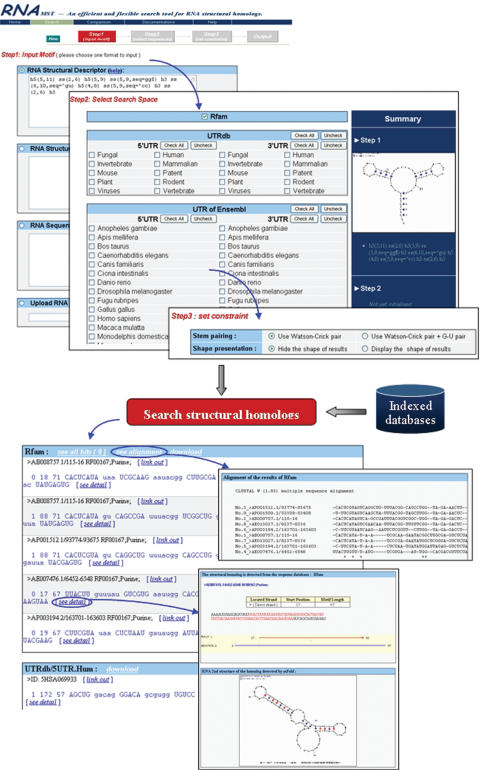
Indexed databases are provided on the web server, including UTRdb (20), Rfam (18), UniGene and UTR sequences from Ensembl. Users can input the description of a desired RNA structural motif via four different ways and select the indexed RNA databases through the user web interface. Moreover, the search results can rapidly be displayed on the webpage. Figure 4 shows the web interface of RNAMST. RNAMST involves the following steps in searching for homologs of a specific RNA structural motif.
Figure 4.
Web interface of RNAMST.
Firstly, users can input the specific RNA structural motif using an RNA structural descriptor. In addition to RNAMST web server also accepts other inputs, including bracket notation, files in ct format and FASTA format RNA sequences.
Secondly, owing to many of the functional RNA structures being discovered in the UTR, RNAMST web server provides UTR related databases, including UTRdb and UTR sequences from Ensembl. Furthermore, RNAMST also provides RNA sequence databases, including Rfam and UniGene. All of these databases were indexed during the preprocessing phase. Users simply need to select the sequences of interest. Accordingly, users can conveniently and rapidly search these databases for homologs of an input RNA structural motif.
Thirdly, the predictive constraints can be specified by users. Besides the input of the structural descriptor, the other formats can set the search similarity constraint to add the mispairs and bulges for the query structural motif. RNAMST supports two kinds of base pairing to fold helices: Watson–Crick pairing (AT and GC pairing), and both Watson–Crick pairing and G–U pairing.
Finally, the RNAMST generates the output for the discovered RNA structural homologs, as well as provides additional information for further analysis, including description of the search sequence containing the homologs, alignment and RNA secondary structure of the homolog.
DISCUSSIONS AND CONCLUSION
RNAMST markedly improves the search time compared with other RNA motif search tools. Although other RNA motif tools such as Palingol, RNAMotif, RNABOB and PatSearch can achieve acceptable search times for searching for strict motifs in a small search sequence, they are not efficient enough to be implemented as a web server when searching in a large search sequence. Notably, the performance comparison among the RNAMotif, RNABOB and our RNAMST is probably unfair due to the RNAMST incorporates a preprocessing strategy to improve the search. In order to provide a convenient web server for identifying RNA structural homologs, the authors suggest that the proposed methodology is a reasonable way to make the web service of this analysis become possible.
Several limitations of current implementation of the RNAMST are addressed below. RNAMST pre-calculates and stores all possible hairpin structures of the search sequence databases. Presently, RNAMST accumulates the hairpin structures which the length of hairpin stem ranges from 3 to 30 nt and the length of hairpin loop ranges from 3 nt to two times the length of the corresponding hairpin stem. Thus, the desired RNA structural homologs beyond the limitation of the pre-indexed database cannot be found. Another limitation is the pre-indexed hairpin structures and the resulting RNA structural homologs are computationally predicted by mfold. Users should note that the predicted RNA secondary structures can provide potential candidates for experimental confirmation. Owing to the complicated structures of the pseodonots, the web server does not support the search for pseudonots yet.
This works developed an efficient and flexible web server, RNAMST, capable of rapidly searching homologs of pre-defined RNA structures against numerous RNA sequences or databases. Table 1 briefly lists the advantages of RNAMST compared with other tools. The web interface enables users to easily implement the various steps of RNAMST. The pre-indexed RNA databases, such as UTRdb, Unigene, UTR of Ensembl and Rfam, are created in advance and supported on the web. The proposed RNAMST web server can provide a fast and convenient analysis for investigators who are interested in regulatory RNA motifs and elements.
Table 1.
Features of RNAMST compared with other tools
| Comparing Features | RNAMST | RNAMotif (14) | RNABOB | PatSearch (15) | Palingol (13) | Riboswitch finder (12) | tRNAscan-SE (11) |
|---|---|---|---|---|---|---|---|
| Diverse RNA structures search | Yes | Yes | Yes | Yes | Yes | — | — |
| High speed search against large amount of sequences | Yes | — | — | — | — | Yes | Yes |
| Web interface with real-time response | Yes | — | — | — | — | Yes | Yes |
| Flexible search with bugle and mispair | Yes | — | — | — | — | — | — |
| Accept multiple RNA description | Yes | — | — | — | — | — | — |
AVAILABILITY
The RNAMST web server will be continuously maintained and updated. The web server is now freely available at http://bioinfo.csie.ncu.edu.tw/~rnamst/.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
Acknowledgments
The authors would like to thank the National Science Council of the Republic of China for financially supporting this research under Contract No. NSC 95-3112-E-009-002. Special thanks for the financially supports from National Research Program For Genomic Medicine (NRPGM), Taiwan. This work was also partially supported by MOE ATU. Funding to pay the Open Access publication charges for this article was provided by National Science Council of the Republic of China.
Conflict of interest statement. None declared.
REFERENCES
- 1.Klaff P., Riesner D., Steger G. RNA structure and the regulation of gene expression. Plant Mol. Biol. 1996;32:89–106. doi: 10.1007/BF00039379. [DOI] [PubMed] [Google Scholar]
- 2.Gygi S.P., Rochon Y., Franza B.R., Aebersold R. Correlation between protein and mRNA abundance in yeast. Mol. Cell. Biol. 1999;19:1720–1730. doi: 10.1128/mcb.19.3.1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gray N.K., Hentze M.W. Regulation of protein synthesis by mRNA structure. Mol. Biol. Rep. 1994;19:195–200. doi: 10.1007/BF00986961. [DOI] [PubMed] [Google Scholar]
- 4.Barrick J.E., Corbino K.A., Winkler W.C., Nahvi A., Mandal M., Collins J., Lee M., Roth A., Sudarsan N., Jona I., et al. New RNA motifs suggest an expanded scope for riboswitches in bacterial genetic control. Proc. Natl Acad. Sci. USA. 2004;101:6421–6426. doi: 10.1073/pnas.0308014101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mandal M., Boese B., Barrick J.E., Winkler W.C., Breaker R.R. Riboswitches control fundamental biochemical pathways in Bacillus subtilis and other bacteria. Cell. 2003;113:577–586. doi: 10.1016/s0092-8674(03)00391-x. [DOI] [PubMed] [Google Scholar]
- 6.Soukup J.K., Soukup G.A. Riboswitches exert genetic control through metabolite-induced conformational change. Curr. Opin. Struct. Biol. 2004;14:344–349. doi: 10.1016/j.sbi.2004.04.007. [DOI] [PubMed] [Google Scholar]
- 7.Winkler W.C., Breaker R.R. Genetic control by metabolite-binding riboswitches. Chembiochem. 2003;4:1024–1032. doi: 10.1002/cbic.200300685. [DOI] [PubMed] [Google Scholar]
- 8.Mignone F., Gissi C., Liuni S., Pesole G. Untranslated regions of mRNAs. Genome Biol. 2002;3:REVIEWS0004. doi: 10.1186/gb-2002-3-3-reviews0004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Eddy S.R. Computational genomics of noncoding RNA genes. Cell. 2002;109:137–140. doi: 10.1016/s0092-8674(02)00727-4. [DOI] [PubMed] [Google Scholar]
- 10.Gray N.K., Wickens M. Control of translation initiation in animals. Annu. Rev. Cell Dev. Biol. 1998;14:399–458. doi: 10.1146/annurev.cellbio.14.1.399. [DOI] [PubMed] [Google Scholar]
- 11.Schattner P., Brooks A.N., Lowe T.M. The tRNAscan-SE, snoscan and snoGPS web servers for the detection of tRNAs and snoRNAs. Nucleic Acids Res. 2005;33:W686–689. doi: 10.1093/nar/gki366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bengert P., Dandekar T. Riboswitch finder--a tool for identification of riboswitch RNAs. Nucleic Acids Res. 2004;32:W154–W159. doi: 10.1093/nar/gkh352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Billoud B., Kontic M., Viari A. Palingol: a declarative programming language to describe nucleic acids' secondary structures and to scan sequence database. Nucleic Acids Res. 1996;24:1395–1403. doi: 10.1093/nar/24.8.1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Macke T.J., Ecker D.J., Gutell R.R., Gautheret D., Case D.A., Sampath R. RNAMotif, an RNA secondary structure definition and search algorithm. Nucleic Acids Res. 2001;29:4724–4735. doi: 10.1093/nar/29.22.4724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Grillo G., Licciulli F., Liuni S., Sbisa E., Pesole G. PatSearch: a program for the detection of patterns and structural motifs in nucleotide sequences. Nucleic Acids Res. 2003;31:3608–3612. doi: 10.1093/nar/gkg548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Benson G. Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Res. 1999;27:573–580. doi: 10.1093/nar/27.2.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Griffiths-Jones S., Moxon S., Marshall M., Khanna A., Eddy S.R., Bateman A. Rfam: annotating non-coding RNAs in complete genomes. Nucleic Acids Res. 2005;33:D121–D124. doi: 10.1093/nar/gki081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hentze M.W., Kuhn L.C. Molecular control of vertebrate iron metabolism: mRNA-based regulatory circuits operated by iron, nitric oxide, and oxidative stress. Proc. Natl Acad. Sci. USA. 1996;93:8175–8182. doi: 10.1073/pnas.93.16.8175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mignone F., Grillo G., Licciulli F., Iacono M., Liuni S., Kersey P.J., Duarte J., Saccone C., Pesole G. UTRdb and UTRsite: a collection of sequences and regulatory motifs of the untranslated regions of eukaryotic mRNAs. Nucleic Acids Res. 2005;33:D141–D146. doi: 10.1093/nar/gki021. [DOI] [PMC free article] [PubMed] [Google Scholar]