Abstract
DISULFIND is a server for predicting the disulfide bonding state of cysteines and their disulfide connectivity starting from sequence alone. Optionally, disulfide connectivity can be predicted from sequence and a bonding state assignment given as input. The output is a simple visualization of the assigned bonding state (with confidence degrees) and the most likely connectivity patterns. The server is available at http://disulfind.dsi.unifi.it/.
INTRODUCTION
Disulfide bridges play a major role in the stabilization of the folding process and, consequently, in studies related to structural and functional properties of specific proteins. In addition, knowledge about the disulfide bonding state of cysteines may help the experimental structure determination process and may be useful in other genomic annotation tasks.
DISULFIND uses a combination of machine learning algorithms to predict intrachain bridges from sequence alone. Similar to many other tools of this kind, it solves the prediction problem in two steps. First, the disulfide bonding state of each cysteine is predicted by a binary classifier; second, cysteines that are known to participate in the formation of bridges are paired to obtain a connectivity pattern.
RELATED WORKS
Early work on bonding state employed representations based on local-window multiple alignment profiles and neural networks for discrimination (1,2). Mucchielli–Giorgi et al. (3) introduced the idea of adding a global descriptor to improve prediction accuracy. Ceroni et al. (4) proposed a method based on a combination of string and vector kernels in conjunction with support vector machines (SVMs). Song et al. (5) applied a linear discriminant using dipeptides as features. Martelli et al. (6) suggested the use of hidden Markov models to refine local predictions obtained via neural networks. SVMs are also used in the method presented in (7).
Prediction of connectivity patterns was pioneered in (8) with a method based on weighted graph matching, implemented in the prediction server DCON. Vullo and Frasconi (9) introduced the use of multiple alignment profiles by means of recursive neural networks (RNNs). In this approach, (that still underpins DISULFIND) a global score is assigned to an entire connectivity pattern. In the DAG RNN approach described in (7,10), the probability for a disulfide bond is computed for each pair of cysteines. The associated DIpro server (which also predicts bonding state) is described in (11). Taskar et al. (12) formulated disulfide connectivity as a structured-output prediction problem and solved it using a generalized large-margin machine. Ferrè and Clote (13) proposed a feedforward neural-network architecture with hidden units associated with cysteine pairs and inputs encoding secondary structure; the method is behind the prediction server DiANNA (14). Zhao et al. (15) confirmed that the profile of distances between bonded cysteines is an important feature for prediction of connectivity patterns. This idea has been further exploited in conjunction with SVMs to develop the method behind the prediction server PreCys (16). Finally, CysView (17) is a server that predict patterns by comparison of a query sequence to annotated data bases.
MATERIALS AND METHODS
Multiple alignment profiles
Prediction of protein structural properties is typically more accurate when incorporating evolutionary information encoded in multiple alignment profiles. Profiles are used in DISULFIND both in bonding state and connectivity prediction. They are calculated by using one iteration of the PSI-BLAST program run on Swiss-Prot and TrEMBL using the BLOSUM62 matrix and an E-value cutoff of 0.005.
Prediction of disulfide bonding state
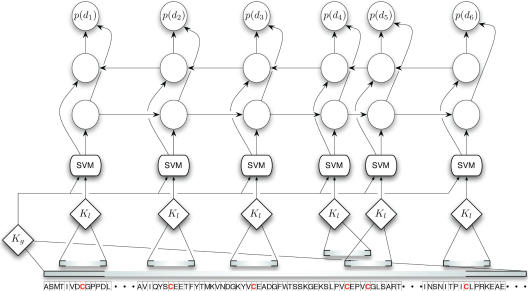
DISULFIND employs an SVM binary classifier to predict the bonding state of each cysteine, followed by a refinement stage that classifies all the cysteines in a chain in a collective fashion (18), that is, by deciding the overall bonding state assignment of an entire chain rather than making several independent predictions (one for each cysteine). The overall architecture is shown in Figure 1. The SVM receives as input both local and global features [see also (3)]. Local features consist of a window of position specific conservations derived from multiple alignment, centered around the target residue. Global features (amino acid composition, chain length, number of cysteines and average cysteine conservation) provide information about the bonding class of the entire chain (all cysteines bonded, none or mix), which is strongly correlated with the subcellular compartment where the protein resides (reducing versus oxidizing environments).
Figure 1.
Architecture of the bonding state predictor. The lower level provides independent cysteine predictions based on a local kernel kl on local attributes, and a global kernel kg on the entire sequence. The upper level is a BRNN (represented here schematically by its graphical model) that outputs a disulfide-bonding probability p(di) for each cysteine, based on all SVM predictions.
The refinement stage is motivated by the observation that single cysteines are not independently sampled. Linkage occurs between pairs forming a disulfide bridge but also among sets of cysteines that coordinate a metal ion. A second source of linkage is due to the fact that bonding state is very often a global property of the protein chain and not a local property of individual cysteines (2,3). The effects of correlation are mitigated in two ways. First, we trained a bidirectional recurrent neural network (BRNN) (19) to predict a globally correct sequence of bonding state assignments, given a (possibly incorrect) sequence of locally calculated predictions. At each cysteine position i, the BRNN output is computed using the logistic function and can be therefore interpreted as the conditional probability p(di) that the cysteine is disulfide-bonded given the input sequence. A position-specific prediction confidence is then defined as
| 1 |
Second, we enforce the number of bonded cysteines to be even (interchain bridges are ignored) using a finite state automaton (shown in Figure 2). Given the sequence of bonding state probabilities (computed by the BRNN), the most likely sequence of bonding states is obtained by running a Viterbi algorithm. Similar ideas (but using a hidden Markov models rather than an automaton) were presented in (6).
Figure 2.
Finite state automaton used in the final stage of bonding state prediction.
Prediction of disulfide connectivity
We assume in this subsection that disulfide-bonding state of cysteines is given (either entered manually by the user or predicted using the method described above). The method used in DISULFIND is fully detailed in (9) and briefly summarized here.
A connectivity pattern can be conveniently represented as an undirected graph whose vertices are cysteines and edges are disulfide bridges. The problem thus consists of mapping an input sequence with annotated cysteines into an output graphs representing disulfide connectivity. This structured output prediction problem can be cast in the traditional supervised learning setting by introducing a regression problem defined as follows. The input is formed by the annotated sequence and a candidate connectivity pattern. The target is a real valued score, defined as the fraction of correctly assigned bridges. During training the target score is known and we use it to train a recursive neural network in regression mode. Prediction is carried out by running the trained network on all possible connectivity patterns and choosing the one yielding maximum score. The number of possible disulfide patterns connecting 2B cysteines is (2B−1)!! where the double factorial n!! is defined as the product of all odd integers that are less or equal to n.
In order to limit computational efforts, DISULFIND can assign at most five disulfide bridges (in this case the number of candidates to be evaluated is 945). Two remarks are relevant to this limitation. First, chains with more than five bridges are rare (no more than 10% of the Swiss-Prot chains annotated with disulfide bridges). Second, the prediction accuracy is already lowfor chains having five bridges because of a limited number of available training examples; hence prediction of patterns with six or more bridges would be very inaccurate.
IMPLEMENTATION
DISULFINDis available both as a standalone service at http://disulfind.dsi.unifi.it/ and as part of PredictProtein (20). The current version (DISULFIND 1.1, released in February 2006), incorporates some improvements in the presentation interface.
Interface
The input to the predictor is entered via an HTTP form using the SEND method. The main fields (see Figure 3) are the following.
Figure 3.
Screenshot of the DISULFIND input form.
Email address The address were results will be sent if the email output option is selected.
Query name An optional field that allows to label the sequence with an user provided identifier.
Amino acid sequence The protein sequence using standard amino acid one-letter codes. Spaces and newlines are automatically stripped.
Predict options In its normal behavior, DISULFIND predicts both bonding state and connectivity. If the bonding state is known in advance, users may check the corresponding option in the user interface and after the form is submitted they will be presented a screen where the bonding state of each cysteine can be manually assigned. In this case only predicted connectivity will be returned.
Output options There are two possible output operation modes. In email mode, after the form is submitted, a job is scheduled in the server and results are returned in ASCII format to the indicated email address. In browser mode, results are returned to the HTTP client (see Figure 4).
Figure 4.
DISULFIND output.
Alternatives By default DISULFIND only returns the most likely connectivity pattern. By setting the number of alternatives to an integer k in the range (1,3), the k best ranking patterns will be returned.
The output presented to the user consists of the original sequence annotated with predictions as shown in Figure 4. The following items are returned in the output screen:
AA The original amino acid sequence;
DB_state Predicted disulfide bonding state (1 = disulfide bonded, 0 = not disulfide bonded);
DB_conf Confidence of disulfide bonding state prediction (0 = low to 9 = high); a red color means that the Viterbi aligner overruled the SVM predition for that residue in order to achieve a consistent prediction at the chain level (i.e. an even number of disulfide bonded cysteines, as interchain bonds are ignored);
Conn_conf Confidence of connectivity assignment given the predicted disulfide bonding state. The confidence in this case is the predicted score associated with the connectivity pattern, i.e. the fraction of correcly assigned bridges—see details in (9). Although the score is a number in (0,1), it should not be confused with the probability that the pattern is correct.
The above output is repeated if multiple alternative patterns are requested. Since all the alternatives share the same bonding state prediction, fields DB_state and DB_conf are only shown in the output presentation of the most likely pattern.
Performance
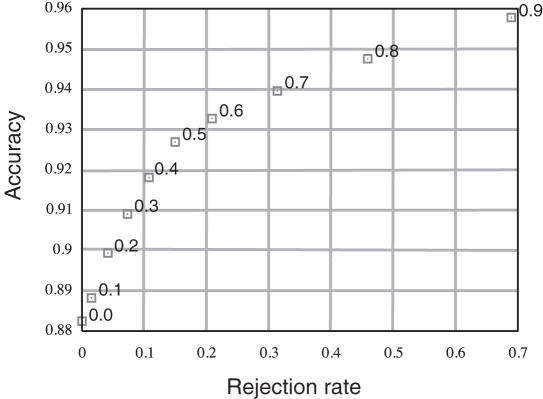
Under regular load conditions, a query can be answered in about 30–60 s. CPU time depends on the sequence length and the number of disulfide bridges. Most CPU time is used by PSI-BLAST for calculation of multiple alignment profiles. The 20-fold cross validation performance of the bonding state prediction stage is reported in Table 1. In order to assess the significance of the confidence score (see Equation 1), we report in Figure 5 the accuracy and rejection rate of the bonding state classifier that abstains when the confidence is lower or equal to a given cutoff. It can be seen, for example, that accuracy improves to Q2 = 92.7% at a rejection rate of 15.0% for a confidence cutoff of 0.5.
Table 1.
DISULFIND bonding state predictor: experimental results on a 20-fold cross validation procedure (PDB Select July 2005)
| Method | Q2 | Qp |
|---|---|---|
| Loc29 | 86 ± 1 | 73 ± 2 |
| BRNN Loc29+f | 88 ± 1 | 82 ± 2 |
| BRNN Loc29+f FSA | 88 ± 1 | 83 ± 2 |
Figure 5.
Accuracy versus rejection rate of the abstaining bonding state predictor for different confidence cutoff values (the rejection rate is the fraction of cysteines that are predicted at a confidence level below the cutoff value shown at the right of each point in the curve).
Concerning disulfide connectivity, leave-one-out estimates of prediction accuracy on a set of 446 Swiss-Prot Sequences (9) are reported in Table 2 [note that results reported in (9) were based on a 4-fold cross validation]. Qp is the fraction of correctly assigned patterns, while Qc is the fraction of correctly predicted bridges. If multiple alternative are selected, the probability that a correct pattern is included increases. Results obtained considering the top k = 3 configurations are Qp = 66.3, Qc = 69.5.
Table 2.
Leave-one-out validation results of disulphide connectity prediction
| Number of bridges | Number of chains | Qp | Qc |
|---|---|---|---|
| 2 | 156 | 75.0 | 75.0 |
| 3 | 146 | 46.6 | 55.7 |
| 4 | 99 | 50.5 | 63.4 |
| 5 | 45 | 17.8 | 42.7 |
| All | 446 | 54.5 | 60.2 |
Statistics
DISULFIND has served a total of over 7000 tasks from almost 50 national domains since April 2003 and is currently serving an average of 60 queries per week. Hundreds of queries per month have been served via PredictProtein since July 2004.
Acknowledgments
The authors would like to thank Rita Casadio and Piero Fariselli for useful discussions and the anonymous reviewers for their suggestions. This research is supported by EU STREP APrIL II (contract no. FP6-508861) and EU NoE BIOPATTERN (contract no. FP6-508803). The work of A.V. is supported by an Embark Fellowship from the Irish Research Council for Science, Engineering and Technology. The Open Access publication charges for this article were waived by Oxford University Press.
Conflict of interest statement. None declared.
REFERENCES
- 1.Fariselli P., Riccobelli P., Casadio R. Role of evolutionary information in predicting the disulfide-bonding state of cysteine in proteins. Proteins. 1999;36:340–346. [PubMed] [Google Scholar]
- 2.Fiser A., Simon I. Predicting the oxidation state of cysteines by multiple sequence alignment. Bioinformatics. 2000;16:251–256. doi: 10.1093/bioinformatics/16.3.251. [DOI] [PubMed] [Google Scholar]
- 3.Mucchielli-Giorgi M.H., Hazout S., Tuffèry P. Predicting the disulfide bonding state of cysteines using protein descriptors. Proteins. 2000;46:243–249. doi: 10.1002/prot.10047. [DOI] [PubMed] [Google Scholar]
- 4.Ceroni A., Frasconi P., Passerini A., Vullo A. Predicting the disulfide bonding state of cysteines with combinations of kernel machines. J. VLSI Signal Processing. 2003;35:287–295. [Google Scholar]
- 5.Song J.-N., Wang M.-L., Li W.-J., Xu W.-B. Prediction of the disulfide-bonding state of cysteines in proteins based on dipeptide composition. Biochem. Biophys. Res. Commun. 2004;318:142–147. doi: 10.1016/j.bbrc.2004.03.189. [DOI] [PubMed] [Google Scholar]
- 6.Martelli P.L., Fariselli P., Casadio R. Prediction of disulfidebonded cysteines in proteomes with a hidden neural network. Proteomics. 2004;4:1665–1671. doi: 10.1002/pmic.200300745. [DOI] [PubMed] [Google Scholar]
- 7.Cheng J., Saigo H., Baldi P. Large-scale prediction of disulphide bridges using kernel methods, two-dimensional recursive neural networks, and weighted graph matching. Proteins. 2006;62:617–629. doi: 10.1002/prot.20787. [DOI] [PubMed] [Google Scholar]
- 8.Fariselli P., Casadio R. Prediction of disulfide connectivity in proteins. Bioinformatics. 2001;17:957–964. doi: 10.1093/bioinformatics/17.10.957. [DOI] [PubMed] [Google Scholar]
- 9.Vullo A., Frasconi P. Disulfide connectivity prediction using recursive neural networks and evolutionary information. Bioinformatics. 2004;20:653–659. doi: 10.1093/bioinformatics/btg463. [DOI] [PubMed] [Google Scholar]
- 10.Baldi P., Cheng J., Vullo A. Large-scale prediction of disulphide bond connectivity. In: Saul L.K., Weiss Y., Bottou L., editors. Advances in Neural Information Processing Systems 17; Cambridge, MA: MIT Press; 2005. pp. 97–104. [Google Scholar]
- 11.Cheng J., Randall A.Z., Sweredoski M.J., Baldi P. SCRATCH: a protein structure and structural feature prediction server. Nucleic Acids Res. 2005;33:W72–W76. doi: 10.1093/nar/gki396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Taskar B., Chatalbashev V., Koller D., Guestrin C. Learning structured prediction models: a large margin approach. Proceedings of the Twenty Second International Conference on Machine Learning (ICML05).2005. [Google Scholar]
- 13.Ferrè F., Clote P. Disulfide connectivity prediction using secondary structure information and diresidue frequencies. Bioinformatics. 2005;21:2336–2346. doi: 10.1093/bioinformatics/bti328. [DOI] [PubMed] [Google Scholar]
- 14.Ferrè F., Clote P. DiANNA: a web server for disulfide connectivity prediction. Nucleic Acids Res. 2005;33:W230–W232. doi: 10.1093/nar/gki412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhao E., Liu H.-L., Tsai C.-H., Tsai H.-K., Chan C.L., Kao C.-Y. Cysteine separations profiles on protein sequences infer disulfide connectivity. Bioinformatics. 2005;21:1415–1420. doi: 10.1093/bioinformatics/bti179. [DOI] [PubMed] [Google Scholar]
- 16.Tsai C.-H., Chen B.-J., Chan C.-H., Liu H.-L., Kao C.-Y. Improving disulfide connectivity prediction with sequential distance between oxidized cysteines. Bioinformatics. 2005;21:4416–4419. doi: 10.1093/bioinformatics/bti715. [DOI] [PubMed] [Google Scholar]
- 17.Lenffer J., Lai P., El Mejaber W., Khan A.M., Koh J.L.Y., Tan P.T.J., Seah S.H., Brusic V. CysView: protein classification based on cysteine pairing patterns. Nucleic Acids Res. 2004;32:W350–W355. doi: 10.1093/nar/gkh475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Getoor L., Friedman N., Koller D., Taskar B. Learning probabilistic models of relational structure. Proceedings 18th International Conf. on Machine Learning; Morgan Kaufmann: San Francisco, CA; 2001. pp. 170–177. [Google Scholar]
- 19.Baldi P., Brunak S., Frasconi P., Soda G., Pollastri G. Exploiting the past and the future in protein secondary structure prediction. Bioinformatics. 1999;15:937–946. doi: 10.1093/bioinformatics/15.11.937. [DOI] [PubMed] [Google Scholar]
- 20.Rost B., Yachdav G., Liu J. The PredictProtein server. Nucleic Acids Res. 2004;32:W321–W326. doi: 10.1093/nar/gkh377. [DOI] [PMC free article] [PubMed] [Google Scholar]