Abstract
NeuroPred is a web application designed to predict cleavage sites at basic amino acid locations in neuropeptide precursor sequences. The user can study one amino acid sequence or multiple sequences simultaneously, selecting from several prediction models and optional, user-defined functions. Logistic regression models are trained on experimentally verified or published cleavage data from mollusks, mammals and insects, and amino acid motifs reported to be associated with cleavage. Confidence interval limits of the probabilities of cleavage indicate the precision of the predictions; these predictions are transformed into cleavage or non-cleavage events according to user-defined thresholds. In addition to the precursor sequence, NeuroPred accepts user-specified cleavage information, providing model accuracy statistics based on observed and predicted cleavages. Neuropred also computes the mass of the predicted peptides, including user-selectable post-translational modifications. The resulting mass list aids the discovery and confirmation of new neuropeptides using mass spectrometry techniques. The NeuroPred application, manual, reference manuscripts and training sequences are available at http://neuroproteomics.scs.uiuc.edu/neuropred.html.
INTRODUCTION
Neuropeptides are bioactive peptides that affect the function of almost every central nervous system (1). Neuropeptidomic studies (2–6) characterize neuropeptides using mass spectrometry and provide high-quality, empirical data on actual neuropeptides. However, because the experimental discovery or confirmation of neuropeptides is time and labor intensive, biochemical characterization of an animal's neuropeptide complement is not available for most species. The increasing number of species that have or are being sequenced at the genomic or transcriptomic level has motivated the development of effective and accurate bioinformatics methodologies to predict neuropeptides from sequence information.
A neuropeptide precursor mRNA sequence can be identified from sequence information (7), and the resulting translated protein sequence includes a signal peptide sequence and one or multiple neuropeptides. An extensive and complicated series of enzymatic processing steps, including cleavage by prohormone or proprotein convertases and other post-translational modifications, occur on the translated protein sequence before the active neuropeptides are created. Prohormone convertases are calcium-dependent serine proteases and each has specific cleavage sites associated with the basic amino acids Lys and Arg (8,9). Kexin, furin, and other prohormone convertases, including PC1, PC2, PC4, PACE4, PC5 and PC7, have overlapping cleavage function, and multiple prohormone convertases are also usually present simultaneously (8,9). Multiple prohormone convertases can cleave the same site, and thus, overcome the functional loss of a specific prohormone convertase. Consequently, the prediction of the resulting neuropeptides from sequence information alone can prove challenging.
While the cleavage motifs for furin and kexin have been extensively studied, there is less information for other prohormone convertases. General observations (often termed rules) for cleavage recognition sites have been proposed (8), usually without knowledge of the acting prohormone convertase. However, these observations stem only from motifs that are cleaved; non-cleaved motifs are typically ignored. Thus, many of these observations are made without regard to cleavage status. Southey et al. (10) predicted precursor cleavages in insects, mammals, birds, fish and other species using a Known Motif approach, based on reported cleavage motifs. Although this approach identified most of the known cleavages, it also had a high rate of false positive results (10).
Other approaches to predict neuropeptide cleavage sites include logistic regression [(11,12), B. R. Southey, A. B. Hummon, T. A. Richmond, S. L. Rodriguez-Zas and J. V. Sweedler, manuscript submitted], and the artificial neural network (13) available in the ProP application (http://www.cbs.dtu.dk/services/ProP). Hummon et al. (11) predicted cleavage sites in mollusk (Aplysia californica) precursors using a logistic regression model on combinations of amino acids and locations, and then applied the predictive function to neuropeptide precursors from a range of organisms. This approach was extended to mammalian precursors (12) and to precursors identified from the Apis mellifera and Drosophila melanogaster genomes (B. R. Southey, A. B. Hummon, T. A. Richmond, S. L. Rodriguez-Zas and J. V. Sweedler, manuscript submitted).
NeuroPred provides a unified interface to predict cleavage sites by employing multiple approaches, based on a wide range of precursors and species, as developed by Hummon et al. (11), Amare et al. (12) and Southey et al. [(10), B. R. Southey, A. B. Hummon, T. A. Richmond, S. L. Rodriguez-Zas and J. V. Sweedler, manuscript submitted]. NeuroPred also has the capability to calculate the mass of the neuropeptides resulting from the predicted cleavages. Made widely available as a web-based application, NeuroPred is a comprehensive resource with which to explore neuropeptide precursor processing and aid in the discovery and confirmation of new neuropeptides.
NEUROPRED
NeuroPred was written for the web using Python (http://www.python.org) and can be accessed from http://neuroproteomics.scs.uiuc.edu/neuropred.html. The main purpose of the NeuroPred tool is to predict the cleavage sites of neuropeptide precursors using logistic regression models trained on experimentally verified cleavage information. In addition, model accuracy indicators and neuropeptide masses can be calculated from the predicted cleavage sites.
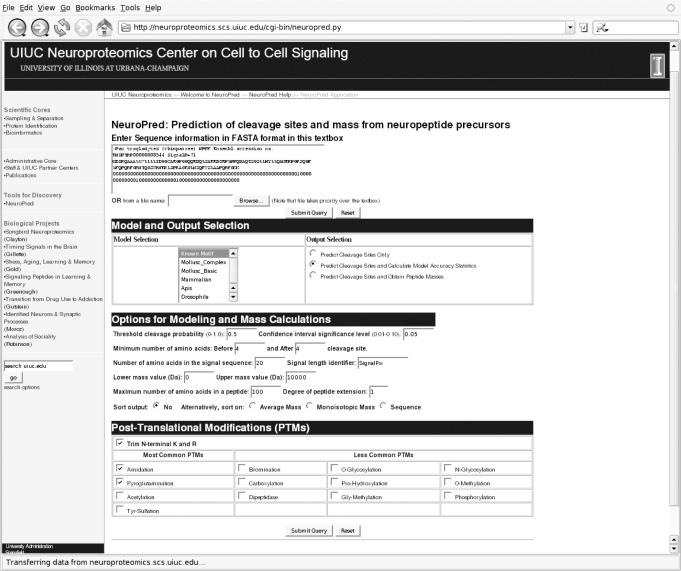
The input required for NeuroPred is one or more sequences, provided in the FASTA format, either entered directly into a text box on the page, or uploaded via a text file. Available user-options include ‘Model and Output Selection’, ‘Options for Modeling and Mass Calculations’ and ‘Post-Translational Modifications’ (Figure 1).
Figure 1.
The NeuroPred input screen with the Chimpanzee NPFF sequence entered.
Model selection
The default model selection is the Known Motif (10); the other models that can be selected are the Mollusk Basic and Mollusk Complex (11), Mammalian (12) and two insect models, one trained using the A.mellifera and the other the D.melanogaster genomic information (B. R. Southey, A. B. Hummon, T. A. Richmond, S. L. Rodriguez-Zas and J. V. Sweedler, manuscript submitted). Multiple models can be used simultaneously to predict cleavage on the same sequence. Specific details about the respective models, including training, sequence information and final terms, are provided by Hummon et al. (11), Amare et al. (12) and Southey et al. [(10), B. R. Southey, A. B. Hummon, T. A. Richmond, S. L. Rodriguez-Zas and J. V. Sweedler, manuscript submitted].
Output selection
Under the default option, ‘Predict Cleavage Sites Only’, NeuroPred will only predict the cleavage sites by calculating the probability of cleavage for each sequence entered for all models selected. This step is completed for all output options. After predicting the cleavage sites, NeuroPred will either compute model accuracy statistics or perform mass calculations, according to the options selected.
Preprocessing
Prior to predicting the probability of cleavage, all precursor sequences undergo a series of preprocessing steps, described in detailed by Southey et al. (B. R. Southey, A. B. Hummon, T. A. Richmond, S. L. Rodriguez-Zas and J. V. Sweedler, manuscript submitted). The size of the window and location of the cleavage site within the window are determined by the model selected; the possibility of cleavage is only considered at a basic site. Preprocessing components that can be changed by the user are the length of the signal peptide and the minimum number of amino acids surrounding the cleavage site. The length of the signal peptide can be specified by the user; this length becomes the global value that is used for all the precursor sequences. Alternatively, the length of the signal peptide can be included in the FASTA label of each sequence, which overrides the global value and may vary across sequences. Examination of a wide range of precursors and the structure of furin (14) suggests that a minimum number of amino acids around the cleavage site are required for cleavage to occur; therefore, the minimum number of amino acids, preceding and following the cleavage site, can also be specified by the user.
Cleavage prediction
Prediction of cleavage is achieved by calculating the probability of cleavage from the logistic regression (15) or Known Motif models for each possible cleavage site. The predicted probability of cleavage is compared with a predefined threshold and converted into cleavage or non-cleavage predictions. A default threshold probability of 50% and a default confidence interval of 95% are displayed for the sites predicted to be cleaved. The asymmetric confidence interval reflects the non-Gaussian nature of the predicted event and parameter space of the probabilities between 0 and 1 (15). Both the threshold probability and confidence interval coverage can be modified by the user.
Output of predicted cleavage
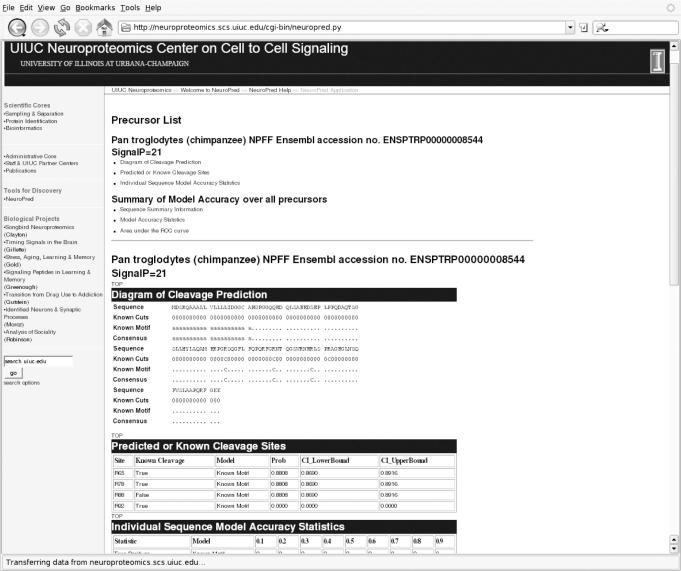
Upon selection of the basic ‘Predict Cleavage Sites Only’ option, the resulting output for each precursor is a diagram that includes the sequence and the predicted cleavage sites (similar to Figure 2). A ‘C’ below amino acid location combinations indicates that the predicted probability of cleavage at that amino acid surpassed the user-defined cleavage probability threshold. The predicted probability of cleavage and associated 95% confidence interval limits are displayed for the amino acid location combinations predicted to be cleaved. The rest of the amino acid location combinations are denoted with a ‘.’ to indicate a non-cleaved prediction. When multiple models are selected, the predicted cleavage sites for each approach are reported on the same diagram, thereby enabling direct comparison of predicted cleavage sites.
Figure 2.
Partial NeuroPred output showing the predicted cleaved and non-cleaved sites and model accuracy statistics for the Chimpanzee NPFF precursor.
Model accuracy statistics
NeuroPred can also assess the performance of the selected approach. By comparing the predicted cleavages with user-entered cleavage information, model accuracy statistics are provided for each individual precursor sequence, and across all precursor sequences entered.
These accuracy statistics are generated by selecting the output option, ‘Predict Cleavage Sites and Calculate Model Accuracy Statistics’. NeuroPred will calculate various model accuracy statistics based on the predicted cleavage sites and user-supplied, or ‘known’, cleavage information. The known cleavage information is entered as a line following the sequence where ‘0’ denotes no cleavage and ‘1’ denotes cleavage. Note that when selecting this output option, if the user-supplied information is not entered, or entered incorrectly, NeuroPred will predict the cleavage sites, but will not provide the accuracy statistics.
The output from the model accuracy statistics selection is similar to that from the ‘Predict Cleavage Sites Only’ option; in addition, the user-entered cleavage information is also represented in the output diagram (Figure 2) to aid in the visualization of correct and incorrect predictions. Cleavage sites that exceed the specified threshold probability are denoted as either true or false predictions. In addition, the probability and confidence interval limits are provided for known cleavage sites that do not exceed the specified threshold probability.
The ‘Predict Cleavage Sites and Calculate Model Accuracy Statistics’ option provides model accuracy statistics for each precursor, and across all precursors, using threshold probabilities ranging from 0.1 to 0.9. For each precursor, model performance is evaluated using the number of predictions of correct cleavage (true positive result), incorrect cleavage (false positive result), correct non-cleavage (true negative result), and incorrect non-cleavage (false negative result) across the length of the precursor. These values are then summarized over all precursors entered. The adequacy of the approach to model cleavage is evaluated across all precursors using the following statistics:
Correct classification rate: number of correctly predicted sites divided by the total number of sites.
Sensitivity (one minus false positive rate): number of true positives divided by the total number of sites cleaved.
Specificity (one minus false negative rate): number of true negatives divided by the total number of sites not cleaved.
Positive predictive power (proportion of sites that are predicted to be cleaved that are true positives): number of true positives divided by the total number of sites predicted to be cleaved.
Negative predictive power (proportion of sites that are not predicted to be cleaved that are true negatives): number of true negatives divided by the total number of sites predicted to not be cleaved.
Correlation coefficient: Mathew's correlation coefficient (16) between observed and predicted cleavage.
Area under the receiver operator characteristic or ROC curve relates sensitivity and 1-specificity (17). Area values lower than 0.7 indicate poor model performance.
Peptide mass prediction
The mass of predicted peptides for multiple precursors can be calculated by NeuroPred, where the masses of the peptides are calculated using common neuropeptide post-translational modifications. For applications using mass spectrometry, a list of the intermediate and final peptides and their associated masses offers more complete information about the precursor-derived peptides than a list of final peptide masses.
The peptides are determined using the model results. Specifically, basic sites are predicted as non-cleaved when there is a consensus from the selected models for non-cleavage. The sequence, after removal of the specified signal peptide for each precursor, is cleaved at the remaining basic sites to provide peptides of different lengths. In order to account for any falsely predicted cleavages and different model predictions, the peptides that are adjacent in the sequence are joined (assuming that cleavage did not occur), and these additional masses are included in the output. This process of joining peptides can be extended up to, but not including, complete sequence after removal of the specified signal peptide.
Options to control the output of the mass prediction include: the number of adjacent sites that can be joined, the range of mass sizes, and the maximum number of amino acids of a predicted peptide resulting from cleavage. The resulting peptides are then further processed using a variety of possible post-translational modifications—with the removal of terminal basic amino acids, amidation and pyroglutamination being selected by default. Other non-default modifications can be selected. The selected post-translational modifications are applied to each peptide where appropriate, resulting in additional peptides. The average and monoisotopic masses are calculated for every peptide.
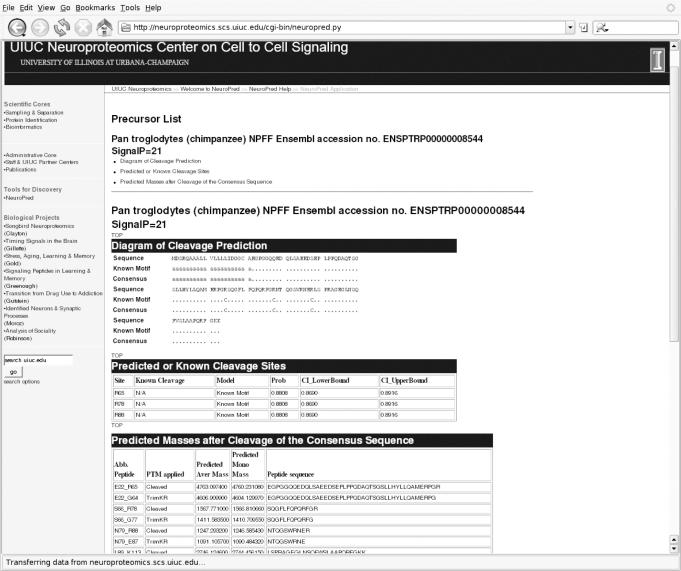
The output (Figure 3) includes a list of peptides and the mass calculation table. The columns of the mass calculation table are the abbreviation of the sequence, the post-translational modifications applied to that sequence, the average and monoisotopic masses, and the full sequence. The user can sort the list of peptides by the combination of actual sequence location of the peptide and post-translational modification, or by either the average or monoisotopic mass.
Figure 3.
Predicted cleavage sites and the mass of predicted peptides after the default post-translation modifications for the Chimpanzee NPFF precursor.
APPLICATION
The non-experimentally confirmed Chimpanzee NPFF precursor sequence reported by Southey et al. (10) is used to demonstrate the use of the NeuroPred application. The FASTA formatted sequence and postulated cleavage information based on the human NPFF sequence are entered in the corresponding text box in NeuroPred. The ‘Predict Cleavage Sites and Calculate Model Accuracy Statistics’ option is selected (Figure 1) and all other settings are at the default values.
The default output is provided for the Chimpanzee NPFF, including the predicted cleavage sites using the default Known Motif approach and the model accuracy statistics (Figure 2). The consensus cleavage sites in the diagram are identical to the cleavage sites predicted by the default model because only one model was selected. The diagram of the cleavage sites indicates that two of the three cleavage sites are correctly identified and one non-cleaved site is incorrectly predicted as cleaved. The probabilities for the false negative site (at position R92) and false positive site (at R88) are 0.89 and 0.0, respectively. The diagram shows that the third amino acid preceding this false positive site is an Arg, thus, fulfilling one of the motifs in the Known Motif model. The false negative site, corresponding to the C-terminal region of the NPAF peptide, occurs because there are no preceding basic amino acids to fulfill any of the motifs in the Known Motif model. This result for the NPFF precursor is consistent with reports of variable location of cleavage across species (10). The model accuracy statistics are presented; however, these statistics are more useful when multiple predictive approaches are selected.
Alternatively, the mass calculations can be obtained by selecting the ‘Predict Cleavage Sites and Obtain Peptide Masses’ option on the input screen. The mass calculations from the Chimpanzee NPFF sequence (Figure 3) show a wide range of possible peptides with different post-translational modifications. The actual Chimpanzee NPFF peptide is expected to have the sequence SQGFLFQPQRFa, based on similarity to the human NPFF (10). One of the peptides generated by cleavage of the NPFF precursor at the predicted cleavage sites is SQGFLFQPQRFGR with a predicted average mass of 1567.77 Da. Enzymes are expected to remove the N-terminal Arg, resulting in a predicted average mass of 1411.58 Da. This peptide ends in Gly and so is likely to be amidated, resulting in the final predicted average mass of 1353.547 Da. This mass is the one most likely to be detected; however, the detection of any of these masses (or MS/MS data) would suggest the presence of this peptide. Thus, the approach implemented in NeuroPred would have correctly predicted the two cleavage sites necessary to generate NPFF from the precursor sequence studied.
AVAILABILITY
The NeuroPred tool, manual, reference publications and the training sequences are available at http://neuroproteomics.scs.uiuc.edu/neuropred.html.
Acknowledgments
This material is based upon work supported by the National Institute of Drug Abuse, through P30 DA018310 to the UIUC Neuroproteomics Center on Cell-to-Cell Signaling, and the National Institutes of Health through GM068946. Funding to pay the Open Access publication charges for this article was provided by NIDA Award No. P30 DA018310.
Conflict of interest statement. None declared.
REFERENCES
- 1.Kandel E.R., Schwartz J.H., Jessell T.M. Principles Of Neural Science, 4th edn. NY: McGraw Hill; 2000. [Google Scholar]
- 2.Svensson M., Skold K., Svenningsson P., Andren P.E. Peptidomics-based discovery of novel neuropeptides. J. Proteome Res. 2003;2:213–219. doi: 10.1021/pr020010u. [DOI] [PubMed] [Google Scholar]
- 3.Haskins W.E., Watson C.J., Cellar N.A., Powell D.H., Kennedy R.T. Discovery and neurochemical screening of peptides in brain extracellular fluid by chemical analysis of in vivo microdialysis samples. Anal. Chem. 2004;76:5523–5533. doi: 10.1021/ac049363y. [DOI] [PubMed] [Google Scholar]
- 4.Baggerman G., Boonen K., Verleyen P., De Loof A., Schoofs L. Peptidomic analysis of the larval Drosophila melangaster central nervous system by two-dimensional capillary liquid cromatograpy quadrupole time-of-flight mass spectrometry. J. Mass Spectrom. 2005;40:250–260. doi: 10.1002/jms.744. [DOI] [PubMed] [Google Scholar]
- 5.Che F.Y., Biswas R., Fricker L.D. Relative quantitation of peptides in wild-type and Cpe(fat/fat) mouse pituitary using stable isotopic tags and mass spectrometry. J. Mass Spectrom. 2005;40:227–237. doi: 10.1002/jms.742. [DOI] [PubMed] [Google Scholar]
- 6.Hummon A.B., Amare A., Sweedler J.V. Discovering new invertebrate neuropeptides using mass spectrometry. Mass Spectrom. Rev. 2006;25:77–98. doi: 10.1002/mas.20055. [DOI] [PubMed] [Google Scholar]
- 7.Hummon A.B., Richmond T.A., Verleyen P., Baggerman G., Huybrechts J., Ewing M.A., Vierstraete E., Rodriguez-Zas S.L., Liliane Schoofs L., Robinson G.E., et al. From the genome to the proteome: uncovering peptides in the Apis brain. Science. 2006 doi: 10.1126/science.1124128. in press. [DOI] [PubMed] [Google Scholar]
- 8.Rockwell N.C., Krysan D.J., Komiyama T., Fuller R.S. Precursor processing by Kex2/Furin Proteases. Chem. Rev. 2002;102:4525–4548. doi: 10.1021/cr010168i. [DOI] [PubMed] [Google Scholar]
- 9.von Eggelkraut R., Beck-Sickinger A.G. Biosynthesis of peptide hormones derived from precursor sequences. Curr. Med. Chem. 2004;11:2651–2665. doi: 10.2174/0929867043364405. [DOI] [PubMed] [Google Scholar]
- 10.Southey B.R., Rodriguez-Zas S.L., Sweedler J.V. Prediction of neuropeptide prohormone cleavages with application to RFamides. Peptides. 2006 doi: 10.1016/j.peptides.2005.07.026. in press. [DOI] [PubMed] [Google Scholar]
- 11.Hummon A.B., Hummon N.P., Corbin R.W., Li L., Vilim F.S., Weiss K.R., Sweedler J.V. From precursor to final peptides: a statistical sequence-based approach to predicting prohormone processing. J. Proteome Res. 2003;2:650–656. doi: 10.1021/pr034046d. [DOI] [PubMed] [Google Scholar]
- 12.Amare A., Hummon A.B., Southey B.R., Zimmerman T.A., Rodriguez-Zas S.L., Sweedler J.V. Bridging neuropeptidomics and genomics with bioinformatics: prediction of mammalian neuropeptide prohormone processing. J. Proteome Res. 2006 doi: 10.1021/pr0504541. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Duckert P., Brunak S., Blom N. Prediction of proprotein convertase cleavage sites. Prot. Eng. Design Sel. 2004;17:107–112. doi: 10.1093/protein/gzh013. [DOI] [PubMed] [Google Scholar]
- 14.Henrich S., Cameron A., Bourenkov G.P., Kiefersauer R., Huber R., Lindberg I., Bode W., Than M.E. The crystal structure of the proprotein processing proteinase furin explains its stringent specificity. Nature Struc. Biol. 2003;10:520–526. doi: 10.1038/nsb941. [DOI] [PubMed] [Google Scholar]
- 15.Collett D. Modelling Binary Data. London: Chapman and Hall; 1991. [Google Scholar]
- 16.Mathews B.W. Comparison of the predicted and observed secondary structure of T4 phage lysozyme. Biochim. Biophys. Acta. 1975;405:442–451. doi: 10.1016/0005-2795(75)90109-9. [DOI] [PubMed] [Google Scholar]
- 17.Green D.M., Swets J.A. Signal Detection Theory And Psychophysics. NY: Wiley; 1966. [Google Scholar]