Abstract
An 840-bp fragment of the 18S rRNA gene was used to identify Cryptosporidium spp. recovered from human immunodeficiency virus (HIV)-infected and -uninfected patients from Kenya, Malawi, Brazil, the United Kingdom, and Vietnam. Initial identification was by Ziehl-Neelsen acid-fast staining. Confirmation was by nested PCR, targeting the most polymorphic region of the 18S rRNA gene. Genotyping was by restriction endonuclease digestion of the PCR product followed by nucleotide sequencing. Among 63 isolates analyzed, four genotypes of Cryptosporidium were identified; 75% of the isolates were of the C. parvum human genotype, while the potentially zoonotic species were of the C. parvum bovine genotype (21.7%), the C. meleagridis genotype (1.6% [one isolate]), and the C. muris genotype (1.6% [one case]). HIV-infected individuals were more likely to have zoonotic genotypes than the HIV-uninfected individuals. Among the C. parvum group, strains clustered distinctly into either human or bovine genotypes regardless of the geographical origin, age, or HIV status of the patients. The intragenotypic variation observed in the C. parvum human genotype was extensive compared to that within the C. parvum bovine genotype group. The variation within genotypes was conserved in all geographical regions regardless of the patients' HIV status. The extensive diversity within genotypes at the 18S rRNA gene locus may limit its application to phylogenetic analyses.
The use of molecular methods in the taxonomy of Cryptosporidium spp. has led to increased recognition of the diversity of the species infecting humans. The main causative agents of human cryptosporidiosis are strains of human and bovine genotypes of the species C. parvum (also called genotype 1 and genotype 2, respectively) (3). Recent reports document human infections with zoonotic species, including C. parvum dog and cat types (now renamed C. canis and C. felis, respectively) and C. meleagridis and C. muris (mainly avian and murine parasites, respectively) (4, 5, 11, 13, 24). A recently recognized Cryptosporidium cervine genotype has been identified in both immunocompromised and immunocompetent people (9). However, the prevalence and significance of the different species and genotypes in both immunocompetent and immunodeficient people are not yet clear. Experimental studies of both humans and animals suggest that diverse species and genotypes show different levels of infectivity and virulence (8). Others have pointed out that zoonotic strains of C. parvum produce more severe infections in humans than the strains found only in humans (7). Moreover, the potential reservoir hosts and transmission pathways for the novel species infecting humans are unclear (3). These issues underscore the importance of the precise identification of species recovered from humans.
So far, a limited number of isolates have been typed from developing countries and especially from human immunodeficiency virus (HIV)-infected people (5, 12, 16, 17, 19, 20, 24), yet cryptosporidiosis is endemic in most tropical regions (2). Extensive genotyping of isolates from different parts of the world is therefore crucial for a more precise mapping of the epidemiology of Cryptosporidium. Our study has targeted the hypervariable region of the 18S rRNA gene in Cryptosporidium for precise parasite identification. Samples analyzed were recovered from children and adult human patients, with or without HIV infection, living in diverse geographical regions. Our study also assessed the extent of variation in sequences from the isolates within each genotype and their effect on the application of this gene target to a phylogenetic analysis of Cryptosporidium.
MATERIALS AND METHODS
Sample collection and DNA extraction.
Cryptosporidium oocysts were recovered from human fecal samples collected from Kenya (32 samples) and stored in 2.5% potassium dichromate at 4°C. Other isolates were from Malawi (11 samples), Vietnam (3 samples), Brazil (7 samples), and the United Kingdom (10 samples; 9 samples from humans and 1 sample from a captive monkey) and were recovered from fecal samples stored at −80°C without preservatives. Specimens stored in potassium dichromate were washed five times in cold distilled water to remove traces of the preservative prior to DNA extraction. A pea size sample of the frozen fecal sample or approximately a 200-μl final suspension of each sample in potassium dichromate was suspended in 200 μl of lysis buffer supplied in a QIAamp kit (QIAGEN Ltd., Crawley, West Sussex, United Kingdom). Oocysts were ruptured by subjecting them to a freeze-thaw cycle of −80°C for 30 min and +80°C for 15 min. DNA was then extracted from the suspension with a QIAamp DNA extraction kit for stool DNA according to the manufacturer's instructions.
Nested-PCR amplification and RFLP analysis.
A highly polymorphic section of the 18S rRNA gene was amplified by nested PCR as described previously (22). The method involves the amplification of an approximately 1,325-bp-long primary product followed by a secondary amplification of an internal fragment with a length of approximately 840 bp. The fragment has been shown to be highly specific for species and genotype identification of Cryptosporidium spp. The different species were thereafter identified by use of restriction digestion of the secondary product. For the restriction digestion (37°C for 1 h), we used 15 μl of the secondary product in a 40-μl (total volume) reaction mixture consisting of 15 U of SspI and 3 μl of restriction buffer for species identification and AsnI (Boehringer Mannheim, Livingston, United Kingdom) at the same concentration for genotyping. The digestion products were separated on a 2% agarose gel and visualized by ethidium bromide staining. Isolates were grouped according to their restriction fragment length polymorphism (RFLP) patterns, and a representative of each group was selected for further analysis.
Sequencing and phylogenetic analysis.
The internal (secondary) fragment of approximately 840 bp was purified with a GenElute commercial kit (Sigma Chemicals, Watford, United Kingdom) according to the manufacturer's instructions. Approximately 1 μg of test DNA was sequenced by a two-directional (reverse and forward) procedure for increased sequence accuracy (ABI automatic sequencer; Lark Technologies, Saffron Walden, United Kingdom). All sequences were subjected to a BLAST search (WUBLAST) to determine their identities and assess their homologies and similarities to those in GenBank. Sequences were aligned with currently published 18S rRNA gene sequences for Cryptosporidium species by using CLUSTALX (EMBL, Heidelberg, Germany) and manual adjustments. Phylogenetic and molecular evolutionary analyses were conducted using MEGA version 2.1 (S. Kumar, K. Tamura, I. B. Jakobsen, and N. Masatoshi, MEGA2: Molecular Evolutionary Genetics Analysis Software, Arizona State University, Tempe, Ariz., 2001). Two thousand replica samplings were analyzed for percent bootstrap values in a neighbor-joining tree. Nucleotide diversity and divergence between C. parvum genotypes were assessed using DnaSP version 3 (14).
Reference sequences retrieved from GenBank were C. parvum human genotype strains HCNV4 (GenBank accession no. AF093489) and HCTX8 (AF159111), C. parvum bovine genotype strain CpBOH (AF093490), C. meleagridis strain CMEL (AF112574), and C. muris K33 (AJ307669), a previously reported strain identified from one of the patients in this study.
Nucleotide sequence accession numbers.
The sequences used in this study have been deposited in the GenBank database under the accession no. AJ493526 to AJ493528, AJ493530 to AJ493542, and AJ493544 to AJ493549.
RESULTS
RFLP analysis of the nested-PCR products showed that 45 (75%) samples were of the C. parvum human genotype and that 16 (21.7%) were of the C. parvum bovine genotype. Single isolates of C. meleagridis (1.6%) and C. muris (1.6%) were also identified. The distribution of the specific genotypes from different geographical areas by patients' age and HIV status is presented in Table 1. The C. parvum human genotype was identified among isolates from each of the geographical regions. The isolate recovered from a captive monkey in the United Kingdom was identified as having a C. parvum human genotype by RFLP analysis, which was confirmed by sequencing.
TABLE 1.
Cryptosporidium genotyping by RFLP analysis
| Source | Species and genotype | HIV statusa | Age | No. of isolates |
|---|---|---|---|---|
| Kenya | C. parvum human | + | Adults | 12 |
| + | Children | 2 | ||
| − | Adults | 9 | ||
| C. parvum bovine | + | Adults | 7 | |
| + | Child | 1 | ||
| C. meleagridis | + | Adult | 1 | |
| C. muris | + | Adult | 1 | |
| Malawi | C. parvum human | + | Child | 1 |
| − | Children | 6 | ||
| C. parvum bovine | + | Child | 1 | |
| − | Children | 3 | ||
| Brazil | C. parvum human | − | Children | 7 |
| Vietnam | C. parvum human | + | Adults | 3 |
| United Kingdom | C. parvum human | NT | Children | 4 |
| C. parvum bovine | NT | Children | 5 | |
| C. parvum human | —b | 1 |
NT, not tested.
—, the isolate derived from a captive monkey.
18S rRNA gene sequence analysis.
The C. meleagridis isolate (recovered from an HIV-infected adult in Kenya) showed 99.8% sequence identity with a C. meleagridis strain retrieved from GenBank (accession no. AF112574), while the C. muris strain showed 100% sequence identity with the published C. muris strain recovered from a rock hyrax. The latter has been documented previously in a case report (4).
In order to assess the extent of the occurrence of heterogeneity in this gene locus within genotypes, further sequence alignment was done, resulting in the selection of 36 isolates that represented the total number of distinct sequences. The remaining sequences showed 100% sequence identity to at least one of the selected isolates.
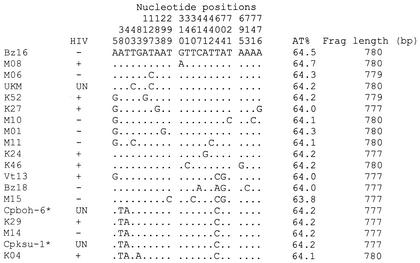
Sequence fragments of about 777 to 780 bp (excluding primer regions) corresponding to positions 224 to 1001 to 1004 of the complete 18S rRNA gene were used in the analysis of nucleotide diversity. Heterogeneity was observed in the 18S rRNA gene for most species, with the highest variation in the C. parvum human genotype occurring in isolates from all regions regardless of their hosts' age or HIV status. Out of the 31 isolates identified as having the C. parvum human genotype, 15 were identified as distinct from three unique types identified from the 10 sequences of isolates identified as belonging to the C. parvum bovine genotype. The extent of the variation within these diverse C. parvum genotype sequences was assessed through DNA polymorphism analysis by using DnaSP (version 3). Within the C. parvum human genotype sequences, 20 polymorphic (segregating) sites with a total of 21 mutations were identified from the entire gene fragment, excluding sites with alignment gaps (780 bp) (Fig. 1 and Table 2). The average number of nucleotide differences per gene fragment was 3.039, while for C. parvum bovine genotype sequences, 10 polymorphic (segregating) sites with a total of 10 mutations were identified in the entire fragment of 777 bp. Within this genotype, the average number of nucleotide differences per gene fragment was 4.6. The results show the high number of mutations present in sequences obtained from each genotype at this gene locus.
FIG. 1.
Variable sites for C. parvum human and bovine genotypes within the 777- to 780-bp hypervariable regions of the 18S rRNA genes. Positions indicated are based on all of the C. parvum sequence alignments noted in Fig. 2, with the BZ16 isolate being considered the consensus (this isolate had 100% sequence identity with the C. parvum human genotype strain HCNV4 [GenBank accession no. AF093489], aligning from positions 224 to 1003 of the 18S rRNA gene, excluding gaps). Sequences shown have been deposited in GenBank with the accession numbers noted in Materials and Methods. Frag, fragment; UN, unknown; ∗, unpublished strains.
TABLE 2.
Analysis of polymorphisms in the 18S rRNA genes of C. parvum human and bovine genotypesa
| Species and genotype | No. of sequences | Total no. of sites | S | Eta | Eta(s) | Theta/eta(s) | Pi | Theta/site | k | Theta/sequence | Tajima's D value |
|---|---|---|---|---|---|---|---|---|---|---|---|
| C. parvum human | 31 | 774 | 20 | 21 | 17 | 16.227 | 0.00393 | 0.00744 | 3.039 | 6.03 | −1.8611 |
| C. parvum bovine | 10 | 774 | 10 | 10 | 4 | 3.000 | 0.00594 | 0.00620 | 4.600 | 4.80 | −0.2982 |
Pi, nucleotide diversity; eta, number of mutations; eta(s), total number of singleton mutations; k, average number of nucleotide pairwise differences; S, number of polymorphic (segregating) sites; eta, total number of mutations; theta, mutation rate per site or per sequence; Tajima's D value, chance that mutations are neutral.
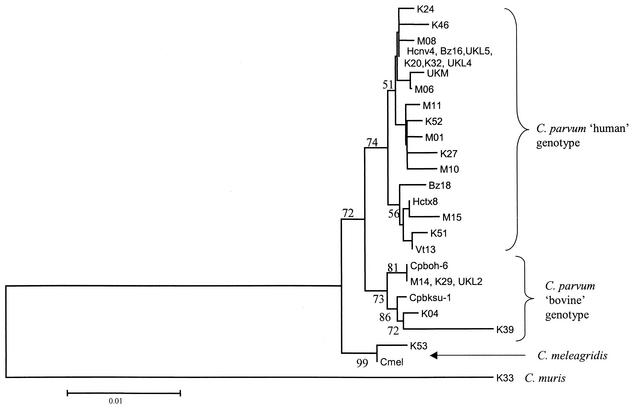
Phylogenetic relationships of the isolates from the different geographical regions were assessed by analysis using the neighbor-joining tree (Fig. 2). Among the 45 sequences identified as belonging to the C. parvum human genotype, 29 (65%) clustered with strain HCNV4. These comprised isolates from all the five geographical regions and included the isolate from a captive monkey in the United Kingdom. The monkey isolate was closest to a human genotype isolate from Malawi (M06). The remaining 16 (35%) human genotype isolates, again occurring in all geographical areas, clustered in another subgroup that was more related to strain HCTX8.
FIG. 2.
Results of a phylogenetic analysis of selected Cryptosporidium isolates from different regions shown in a neighbor-joining tree. K, Kenya; M, Malawi; Bz, Brazil; Vt, Vietnam; UKL, United Kingdom (human); UKM, United Kingdom monkey isolate. Other labels are per published strain identifications. Bootstrap values greater than 50% are shown.
DISCUSSION
The C. parvum human genotype was the most prevalent, occurring in over 75% of all isolates from both HIV-infected and -uninfected people. It was the only genotype identified in isolates from Vietnam and Brazil. However, 22.2% of the potentially zoonotic genotypes were identified in the 29 HIV-infected people compared to 11.1% in the 25 people who were HIV uninfected, but the difference was not statistically significant (P = 0.4). Our report on the occurrence of diverse genotypes infecting humans is in agreement with the published literature on human cryptosporidiosis (5, 15, 17, 24). The presence of eight C. parvum bovine genotype isolates from Kenya mostly from HIV-infected persons is significant. All C. parvum human isolates previously identified from the region have been of the human genotype (5, 12). The C. parvum human genotype is thought to be the most prevalent in most parts of the world except in some parts of the United Kingdom (23). C. meleagridis has been identified previously in Kenyan isolates also from HIV-infected people (5, 12). Our study together with previous reports shows that infection with potentially zoonotic Cryptosporidium species may be widespread in the region.
There are no current reports, to the best of our knowledge, on the prevalence of specific genotypes in Malawi or Vietnam, while one previous report has identified the C. parvum human genotype in isolates from Brazil (10). While most isolates in our study from these regions were identified as belonging to the C. parvum human genotype, the number was small and the possibility that zoonotic genotypes exist cannot be ruled out. Four cases of infection with isolates of the C. parvum bovine genotype were identified in children from Malawi, one of which was an HIV-infected child. It is therefore possible that strains with other genotypes infect humans in these regions. Recent reports from Thailand and Peru show the occurrence of C. muris, C. felis, and C. meleagridis in children (17, 24). The relevance of zoonotic species and different C. parvum genotypes and subgenotypes to specific symptoms in humans and animals has not been fully elucidated. It is known that Cryptosporidium infection in animals and humans produces different levels of infectivity and severity and even different responses to treatment (1, 2, 8). Moreover, the potential host ranges and transmission pathways of the potentially zoonotic species to humans are yet to be documented, as having pets does not appear to be a risk factor (3).
Previous studies of the 18S rRNA gene have shown that the locus can be useful for the specific identification of most Cryptosporidium species (5, 6, 11, 15-17, 19, 20, 24). However, the lack of specific variation in the isolates resulting from either the HIV status of the patient or the geographical origin of the isolate suggests that the locus may not be appropriate for isolate-specific fingerprinting. There has also been extensive application of this gene in the phylogeny of Cryptosporidium, with indications that the presence of heterogenous copies within the genome of a single organism does not affect the overall phylogenetic position of the organism (6, 17, 21). Tests to determine the extent of C. parvum nucleotide diversity show that the heterogenous copies at this gene locus have a high mutation rate that is more pronounced within the C. parvum human genotype. As the results of other studies have demonstrated, our results show that the presence of heterogenous copies of the 18S rRNA gene may result in the overestimation of divergence, especially within C. parvum genotype groups (21). These factors might limit the application of the locus in a fine-scale phylogenetic analysis.
The heterogeneity may also suggest the occurrence of an extensive genetic mix of natural populations of the C. parvum human genotype. Similar mixed profiles of C. parvum strains occurring at a high frequency have been observed in studies using RFLP markers on the β-tubulin gene (18). The findings show the limitations of the use of this gene and indicate the need for further research on the genetic structures of C. parvum based on the 18S rRNA gene. However, the ability to amplify this gene fragment from different species and genotypes of the organism with one set of primers makes this locus the most appropriate for screening where the species and types of Cryptosporidium organisms occurring are unknown.
Acknowledgments
We thank Lihua Xiao of the CDC for supplying positive control samples, Martin Donnely for advice, and Ricardo Gurgel (University of Sergipe, Aracaju, Brazil) and James Sarkodie for assistance in sampling.
This study was partly funded by a Ph.D. research grant to Wangeci Gatei from the Association of Commonwealth Universities at the Liverpool School of Tropical Medicine, Liverpool, United Kingdom.
REFERENCES
- 1.Caccio, S., E. Pinter, R. Fantini, I. Mezzaroma, and E. Pozio. 2002. Human infection with Cryptosporidium felis: case report and literature review. Emerg. Infect. Dis. 8:85-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Casemore, D. P. 1997. Cryptosporidiosis—human and animal epidemiology, p. 64-95. In R. Fayer (ed.), Cryptosporidium and cryptosporidiosis. CRC Press, Boca Raton, Fla.
- 3.Fayer, R., U. Morgan, and S. J. Upton. 2000. Epidemiology of Cryptosporidium: transmission, detection and identification. Int. J. Parasitol. 30:1305-1322. [DOI] [PubMed] [Google Scholar]
- 4.Gatei, W., R. W. Ashford, N. J. Beeching, S. K. Kamwati, J. Greensill, and C. A. Hart. 2002. Cryptosporidium muris infection in an HIV-infected adult, Kenya. Emerg. Infect. Dis. 8:204-206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Morgan, U., R. Weber, L. Xiao, I. Sulaiman, R. C. Thompson, W. Ndiritu, A. Lal, A. Moore, and P. Deplazes. 2000. Molecular characterization of Cryptosporidium isolates obtained from human immunodeficiency virus-infected individuals living in Switzerland, Kenya, and the United States. J. Clin. Microbiol. 38:1180-1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Morgan, U. M., C. C. Constantine, D. A. Forbes, and R. C. Thompson. 1997. Differentiation between human and animal isolates of Cryptosporidium parvum using rDNA sequencing and direct PCR analysis. J. Parasitol. 83:825-830. [PubMed] [Google Scholar]
- 7.Okhuysen, P. C., and C. L. Chappell. 2002. Cryptosporidium virulence determinants—are we there yet? Int. J. Parasitol. 32:517-525. [DOI] [PubMed] [Google Scholar]
- 8.Okhuysen, P. C., C. L. Chappell, J. H. Crabb, C. R. Sterling, and H. L. DuPont. 1999. Virulence of three distinct Cryptosporidium parvum isolates for healthy adults. J. Infect. Dis. 180: 1275-1281. [DOI] [PubMed] [Google Scholar]
- 9.Ong, C. S., D. L. Eisler, A. Alikhani, V. W. Fung, J. Tomblin, W. R. Bowie, and J. L. Isaac-Renton. 2002. Novel Cryptosporidium genotypes in sporadic cryptosporidiosis cases: first report of human infections with a cervine genotype. Emerg. Infect. Dis. 8:263-268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ortega, Y. R., R. R. Sheehy, V. A. Cama, K. K. Oishi, and C. R. Sterling. 1991. Restriction fragment length polymorphism analysis of Cryptosporidium parvum isolates of bovine and human origin. J. Protozool. 38:40S-41S. [PubMed]
- 11.Pedraza-Diaz, S., C. Amar, A. M. Iversen, P. J. Stanley, and J. McLauchlin. 2001. Unusual Cryptosporidium species recovered from human faeces: first description of Cryptosporidium felis and Cryptosporidium ‘dog type' from patients in England. J. Med. Microbiol. 50:293-296. [DOI] [PubMed] [Google Scholar]
- 12.Peng, M. M., O. Matos, W. Gatei, P. Das, M. Stantic-Pavlinic, C. Bern, I. M. Sulaiman, S. Glaberman, A. A. Lal, and L. Xiao. 2001. A comparison of Cryptosporidium subgenotypes from several geographic regions. J. Eukaryot. Microbiol. 48(Suppl.):28S-31S. [DOI] [PubMed]
- 13.Pieniazek, N. J., F. J. Bornay-Llinares, S. B. Slemenda, A. J. da Silva, I. N. Moura, M. J. Arrowood, O. Ditrich, and D. G. Addiss. 1999. New Cryptosporidium genotypes in HIV-infected persons. Emerg. Infect. Dis. 5:444-449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rozas, J., and R. Rozas. 1999. DnaSP version 3: an integrated program for molecular population genetics and molecular evolution analysis. Bioinformatics 15:174-175. [DOI] [PubMed] [Google Scholar]
- 15.Spano, F., L. Putignani, A. Crisanti, P. Sallicandro, U. M. Morgan, S. M. Le Blancq, L. Tchack, S. Tzipori, and G. Widmer. 1998. Multilocus genotypic analysis of Cryptosporidium parvum isolates from different hosts and geographical origins. J. Clin. Microbiol. 36:3255-3259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sulaiman, I. M., L. Xiao, C. Yang, L. Escalante, A. Moore, C. B. Beard, M. J. Arrowood, and A. A. Lal. 1998. Differentiating human from animal isolates of Cryptosporidium parvum. Emerg. Infect. Dis. 4:681-685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tiangtip, R., and S. Jongwutiwes. 2002. Molecular analysis of Cryptosporidium species isolated from HIV-infected patients in Thailand. Trop. Med. Int. Health 7:357-364. [DOI] [PubMed] [Google Scholar]
- 18.Widmer, G., L. Tchack, C. L. Chappell, and S. Tzipori. 1998. Sequence polymorphism in the β-tubulin gene reveals heterogeneous and variable population structures in Cryptosporidium parvum. Appl. Environ. Microbiol. 64:4477-4481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Widmer, G., L. Tchack, F. Spano, and S. Tzipori. 1998. A study of Cryptosporidium parvum genotypes and population structure. Mem. Inst. Oswaldo Cruz 93:685-686. [DOI] [PubMed] [Google Scholar]
- 20.Widmer, G., S. Tzipori, C. J. Fichtenbaum, and J. K. Griffiths. 1998. Genotypic and phenotypic characterization of Cryptosporidium parvum isolates from people with AIDS. J. Infect. Dis. 178:834-840. [DOI] [PubMed] [Google Scholar]
- 21.Xiao, L., J. R. Limor, L. Li, U. Morgan, R. C. Thompson, A. A. Lal. 1999. Presence of heterogeneous copies of the small subunit rRNA gene in Cryptosporidium parvum human and marsupial genotypes and Cryptosporidium felis. J. Eukaryot. Microbiol. 46:44S-45S. [PubMed]
- 22.Xiao, L., U. M. Morgan, J. Limor, A. Escalante, M. Arrowood, W. Shulaw, R. C. A. Thompson, R. Fayer, and A. A. Lal. 1999. Genetic diversity within Cryptosporidium parvum and related Cryptosporidium species. Appl. Environ. Microbiol. 65:3386-3391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xiao, L., U. M. Morgan, R. Fayer, R. C. Thompson, and A. A. Lal. 2000. Cryptosporidium systematics and implications for public health. Parasitol. Today 16:287-292. [DOI] [PubMed] [Google Scholar]
- 24.Xiao, L., C. Bern, J. Limor, I. Sulaiman, J. Roberts, W. Checkley, L. Cabrera, R. H. Gilman, and A. A. Lal. 2001. Identification of 5 types of Cryptosporidium parasites in children in Lima, Peru. J. Infect. Dis. 183:492-497. [DOI] [PubMed] [Google Scholar]