Abstract
In this study a systematic attempt has been made to integrate various approaches in order to predict allergenic proteins with high accuracy. The dataset used for testing and training consists of 578 allergens and 700 non-allergens obtained from A. K. Bjorklund, D. Soeria-Atmadja, A. Zorzet, U. Hammerling and M. G. Gustafsson (2005) Bioinformatics, 21, 39–50. First, we developed methods based on support vector machine using amino acid and dipeptide composition and achieved an accuracy of 85.02 and 84.00%, respectively. Second, a motif-based method has been developed using MEME/MAST software that achieved sensitivity of 93.94 with 33.34% specificity. Third, a database of known IgE epitopes was searched and this predicted allergenic proteins with 17.47% sensitivity at specificity of 98.14%. Fourth, we predicted allergenic proteins by performing BLAST search against allergen representative peptides. Finally hybrid approaches have been developed, which combine two or more than two approaches. The performance of all these algorithms has been evaluated on an independent dataset of 323 allergens and on 101 725 non-allergens obtained from Swiss-Prot. A web server AlgPred has been developed for the predicting allergenic proteins and for mapping IgE epitopes on allergenic proteins (http://www.imtech.res.in/raghava/algpred/). AlgPred is available at www.imtech.res.in/raghava/algpred/.
INTRODUCTION
Allergy involves a series of complex reactions and both intrinsic and extrinsic factors contribute to the development of the disease and triggering of the symptoms. Type I hypersensitive reaction is induced by certain types of antigens referred to as allergens that elicit specific IgE antibodies or from cross-reactivity between common homologous allergens from different sources (1–3). Typical symptoms for this type of allergies are rhinitis, asthma and atopic eczema, but more severe reactions such as acute and fatal anaphylactic shock can also occur. It affects a large population with very high prevalence particularly of skin sensitization (4,5). Most allergic responses occur on mucous membrane surface in response to allergens that enter the body by either inhalations or ingestion.
The prediction of allergenic proteins is becoming very important at present due to the use of modified proteins in foods (genetically modified foods), therapeutics and bio-pharmaceuticals (5–8). World Health Organization (WHO) and the Food and Agriculture Organization (FAO) proposed guidelines to assess the potential allergencity of proteins (9,10). The bioinformatics part of the guidelines of 2001 says that a protein is potentially allergenic if it either has an identity of at least six contiguous amino acids or a minimum of 35% sequence similarity over a window of 80 amino acids when compared with known allergens. The six amino acid identity rule has been shown to produce false positive results (11–17). Stadler and Stadler (17) claimed that the 35% over 80 residues rule might be too conservative since allergenic cross-reactivity typically requires >70% identity across the entire protein (18). In 2003, the Codex Alimentarius Commission (Codex) conveyed a panel of international food safety regulators to review the FAO/WHO 2001 recommendations and recognized the uncertainties associated with different tests. They suggest weight of evidence approach rather than a specific decision tree approach (suggested in FAO/WHO 2001). They recommended various tests for examining allergenic behavior of proteins that includes source of gene, sequence similarities with known allergens, stability of protein and IgE bindings (19).
A number of methods have been developed in past for predicting allergens based on various criteria's that includes (i) motif based approach using MEME/MAST (17); (ii) K-nearest neighbor classifier (20,21) and (iii) similarity search against IgE epitopes, epitope profiles, structure profiles and the like (22). Each approach has their own limitations, e.g. epitopes-based approach fails due to limited number of known epitopes and low accuracy of existing epitope prediction methods (23,24). In this paper we made an attempt to predict the allergenic proteins based on various approaches. First, a standard method has been developed for predicting allergens based on amino acid and dipeptide composition of proteins using support vector machine (SVM). In the second approach, motif-based technique has been used for predicting allergens using the software MEME/MAST. Third, we assigned a protein as allergen, if it has a segment similar to allergen representative proteins (ARPs). In fourth approach, a protein is assigned allergen if it have segment identical to known IgE epitopes.
MATERIALS AND METHODS
Collection and compilation of allergens
Dataset
The dataset used in this study were obtained from http://www.slv.se/templatesSLV/SLV_Page_9343.asp (12), which contains 578 allergens and 700 non-allergens (derived from food). The epitopes were obtained from various sources that include 56 IgE epitopes from Bcipep database (25) and 157 IgE epitopes from SDAP database (22). Finally we got 178 epitopes after removing redundant-epitopes and epitopes having less than five amino acids. These IgE epitopes were scanned against dataset of allergic and non-allergic proteins. In addition to scan at a fix percent of identity (PID), we also scanned at different PID based on length of IgE epitopes.
Dataset partitioning
One of the challenges in developing any prediction method is to minimize the similarity between the proteins used for training and the proteins used for testing. Removing redundancy reduces the number of proteins used for training, which is not good for any learning method. A different approach to minimize similarity between proteins used for training and testing without reducing total number of proteins has been used in this study (26). First proteins were clustered based on similarity using BLAST E-value 8E−4 (26% identity for one sequence pair). These clusters were grouped into five sets in such a way, that each set has nearly equal number of sequences, where all proteins of a given cluster are kept in one set. As sequences in one cluster do not have similarity (E-value of 8E−4) with sequences of other clusters so sequences in one set will not have similarity with sequences of other sets.
Independent dataset
The performance of methods has been evaluated on a blind or independent dataset obtained from Li et al. (15). Initially this dataset have 664 allergens where allergens obtained from various sources that includes 238 allergens from International union of immunological societies (IUIS) (www.allergen.org/List.htm), 270 from Swiss-Prot's Allergen Index (www.expasy.org/cgi-bin/lists?allergen.txt), 1171 from the biotechnology information for food safety database (BIFS) (www.iit.edu/~sgendel/fa.htm) and 752 from food allergy research and resource program (FARRP) (www.allergenonline.com). In this dataset no two sequence have identity >95%. We generate an independent dataset of 323 proteins from the dataset of 664 proteins by removing the common allergens present in the dataset of Bjorklund et al. (12). These 323 proteins were used as independent dataset for evaluating different methods. We also checked the similarity of 323 allergen sequences to 578 sequences used in developing the algorithm using PROSET software (27). It was observed that 48 sequences in independent dataset have 95% or more similarity with protein sequences of 578 used for developing methods.
Swiss-Prot proteins
The performance of various approaches has been evaluated in terms of rate of false prediction on 101 725 non-allergens. These non-allergens extracted from 211 104 entries of Swiss-Prot (on 49.2 release of March 07, 2006) after excluding sequences having ‘Allergy’ in any field or having word ‘Similarity’ or ‘Probably’ in comment field (28). Thus this dataset consists of 101 725 well-annotated non-allergenic proteins and considered as independent dataset of non-allergens (negative examples).
ARPs collection
The dataset of ARPs consists of 2890 ARPs (24 amino acid peptides) obtained from Bjorklund et al. (12). They collected high-quality repositories of amino acid sequences of proteinaceous allergens (allergen database) and non-allergens (consumed commodities, such as rice, apple, tomato and the like) and generated all possible overlapping 24mer peptides for both types of proteins (allergen and non-allergens). Based on the global similarity scores of each allergen peptide, a set containing 2890 ARPs was created which had high similarity in allergenic proteins but not in non-allergenic proteins.
MEME/MAST
MEME/MAST (29,30), version 3.0.4, obtained from http://meme.sdsc.edu/meme/ website. MEME (Multiple Em for Motif Elicitation) is a tool for discovering motifs in a group of related protein sequences. A motif is a sequence pattern that occurs repeatedly in a group of related protein sequences. MEME represents motifs as position-dependent letter-probability matrices, which describe the probability of each possible letter at each position in the pattern. MEME takes as input a group of protein sequences (the training set) and outputs as many motifs as requested. MEME uses statistical modeling techniques to automatically choose the best width, number of occurrences and description for each motif. MAST (Motif Alignment and Search Tool) is a tool for searching biological sequence databases for sequences that contain one or more of a group of known motifs. MAST takes as input a file containing the descriptions of one or more motifs and searches sequence databases that have been created that match the motifs.
Support vector machine
The SVM has been implemented using SVM_light (31), which allow users to select various parameters and various kernel functions like radial basis function (RBF), polynomial. Preliminary tests showed that the RBF kernel gives results better than other kernels. Therefore, in this work we used the RBF kernel for all the experiments. The input vectors used were amino acid composition (20 vectors) and dipeptide composition (400 vectors) of each protein sequence.
Protein features
Amino acid composition. Amino acid composition is the fraction of each amino acid in a protein. The fraction of all 20 natural amino acids was calculated using the following equations:
where i can be any amino acid.
Dipeptide composition. Dipeptide composition was used to encapsulate the global information about each protein sequence, which gives a fixed pattern length of 400 (20 × 20). This representation encompassed the information about amino acid composition along local order of amino acid. The fraction of each dipeptide was calculated using following equation:
where dipep (i) is 1 out of 400 dipeptides.
Fivefold cross-validation
The performance of all methods developed in this study is evaluated using 5-fold cross validation. In 5-fold cross validation the dataset has been divided into five sets, where each set has nearly equal number of allergens and non-allergens. The training and testing of every method has been carried out five times, each time using one distinct set for testing and remaining four sets for training. The overall performance of a method is the average performance over five sets.
Performance measures
A standard set of parameters has been used to evaluate the performance of various methods developed in this study. Following is a brief description of the parameters (32): (i) sensitivity, also referred to as recall, is the percent of correctly predicted allergen epitopes (15); (ii) specificity is the percent of correctly predicted non-allergen epitopes; (iii) accuracy is the proportion of correctly predicted epitopes; (iv) PPV (positive prediction value, also referred to as precision) is the probability of correct positive prediction (15); (v) NPV (negative prediction value) is the probability of correct negative prediction; and (vi) Matthew's correlation coefficient (MCC). The parameters may be calculated by the following equations.
where TP and FN refer to true positive and false negatives and TN and FP refer to true negatives and false positives.
RESULTS
Standard SVM-based method
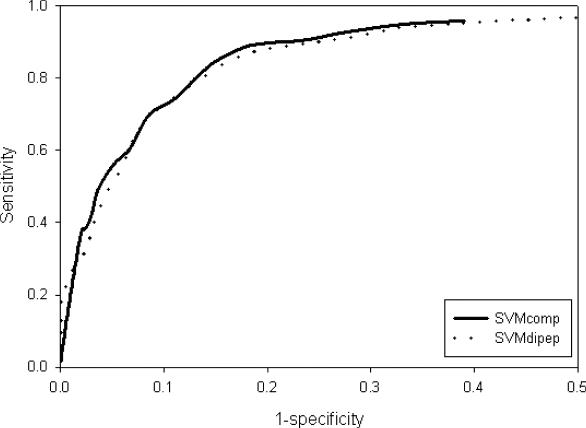
First, amino acid composition of all proteins (allergen and non-allergen) has been computed. Then this composition is used as an input vector of dimension 20 for training and testing of SVM (33–35). The performance of SVM-based methods has been optimized by tuning SVM parameters, in order to achieve maximum accuracy with nearly equal sensitivity and specificity. As shown in Table 1, we achieved accuracy around 85% using this approach. This method, correctly predicted 95% of allergens at specificity of 61%. It also correctly predicted 34% of allergens at specificity around 98 with 94% probability of correct prediction. Similarly, we developed a SVM-based method using dipeptide composition (Supplementary Table S1; http://www.imtech.res.in/raghava/algpred/suppl.html). The performance of the SVM modules based on amino acid and dipeptide composition has been shown in Figure 1. The results indicated that SVM module based on amino acid composition is slightly better than the dipeptide composition. We also compute PPV and NPV values at different threshold value range using SVM module based on amino acid and dipeptide composition and has been shown in Table 2.
Table 1.
Performance of SVM-based method using amino acid composition
| Threshold | Sensitivity | Specificity | Accuracy | PPV | NPV | MCC |
|---|---|---|---|---|---|---|
| 1.0 | 0.3374 | 0.9829 | 0.6918 | 0.9417 | 0.6442 | 0.4336 |
| 0.8 | 0.4243 | 0.9700 | 0.7239 | 0.9208 | 0.6729 | 0.4843 |
| 0.6 | 0.5200 | 0.9586 | 0.7608 | 0.9116 | 0.7093 | 0.5456 |
| 0.4 | 0.5878 | 0.9386 | 0.7804 | 0.8871 | 0.7357 | 0.5732 |
| 0.2 | 0.6539 | 0.9243 | 0.8024 | 0.8765 | 0.7657 | 0.6099 |
| 0.0 | 0.7357 | 0.8929 | 0.8220 | 0.8494 | 0.8054 | 0.6422 |
| −0.2 | 0.8383 | 0.8543 | 0.8471 | 0.8253 | 0.8667 | 0.6930 |
| −0.4 | 0.8887 | 0.8186 | 0.8502 | 0.8009 | 0.9009 | 0.7053 |
| −0.6 | 0.9061 | 0.7614 | 0.8267 | 0.7573 | 0.9096 | 0.6680 |
| −0.8 | 0.9391 | 0.6957 | 0.8055 | 0.7171 | 0.9347 | 0.6441 |
| −1.0 | 0.9583 | 0.6100 | 0.7671 | 0.6687 | 0.9489 | 0.5933 |
The boldface indicates the best result
Figure 1.
The ROC plot of SVM-based method using amino acid (SVMcomp) and dipeptide (SVMdipep) composition.
Table 2.
The probability of correct prediction of allergens and non-allergens is shown in terms of PPV and NPV, respectively, for SVM-based methods using residue and dipeptide composition
| SVM score (range) | SVM based on composition | SVM based on dipeptide composition | ||
|---|---|---|---|---|
| PPV | NPV | PPV | NPV | |
| 1.0–0.8 | 85.64 | 67.96 | 100.00 | 59.74 |
| 0.8–0.6 | 87.05 | 71.53 | 82.97 | 62.40 |
| 0.6–0.4 | 81.83 | 74.03 | 86.55 | 66.47 |
| 0.4–0.2 | 74.81 | 76.94 | 85.88 | 72.01 |
| 0.2–0.0 | 70.05 | 80.74 | 74.14 | 79.04 |
| 0.0 to −0.2 | 64.55 | 86.61 | 63.10 | 85.56 |
| −0.2 to −0.4 | 47.13 | 89.71 | 39.40 | 89.34 |
| −0.4 to −0.6 | 18.21 | 71.24 | 27.66 | 92.40 |
| −0.6 to −0.8 | 22.82 | 92.94 | 13.26 | 74.19 |
| −0.8 to −1.0 | 15.19 | 94.18 | 8.69 | 75.22 |
Presence of IgE epitopes
In this approach, a protein is predicted allergen if it has one or more IgE epitopes. An attempt has been made to develop a method for predicting IgE epitopes but it achieved very poor performance due to complexity and variable length of IgE epitopes (See Supplementary Data, http://www.imtech.res.in/raghava/algpred/suppl.html). Thus in this study, a similarity based approach has been used, whereby a protein is predicted to be an allergen if it has a region/peptide identical to a known IgE epitopes. First all 183 IgE epitopes were scanned against all proteins in the dataset. When a stringent condition of 100% identity was used 61 hits were found among the allergens and 16 hits—in non-allergens. Ideally, IgE epitope should not be present in non-allergens, therefore epitopes present in non-allergens were further analyzed. It has been observed that most of the epitopes founds in non-allergen were short in length (three or four residues); e.g. epitopes HWR, IRRA and YHVP have 7, 6 and 1 hits, respectively, in non-allergens. Owing to their short length they were unable to provide specificity. Thus only those epitopes which have five or more than five residues were studied further (178 epitopes). As shown in Table 3, for PID 100 we got 56 hits in allergen (or 9.69% of allergens were correctly assigned as allergens) and 2 hits in non-allergens (or 0.28% of non-allergens assigned wrongly as allergens). In order to increase the sensitivity or percent coverage of allergens we relaxed the criteria. Although the sensitivity increased, the percent of false assignment of non-allergen to allergen also increased when we relaxed the criteria. Thus, we set PID cut-off based on length of epitope rather than using a uniform cut-off. In this way we were able to achieve better sensitivity without loosing significant specificity. The best results were obtained using PID865 where PID cut-off is 80, 60 and 50 for epitopes having residues <10, between 10 and 15 and >15, respectively.
Table 3.
Searching of 178 IgE epitopes in protein dataset consists of 578 allergens and 700 non-allergens
| Approach | PID (cut-off) | Total hits | |
|---|---|---|---|
| Allergens | Non-allergens | ||
| PID100 | 100 | 56 (9.69%) | 2 (0.28%) |
| PID81 | >80 | 77 (13.32%) | 8 (1.11%) |
| PID80 | ≥80 | 102 (17.65%) | 74 (10.57 %) |
| PID876 | >80 (epitopes have ≤9 amino acids) | 91 (15.74%) | 11 (1.57%) |
| ≥70 (epitopes have >9 and ≤15 amino acids) | |||
| >60 (epitopes have >15 amino acids) | |||
| PID865 | >80 (epitopes have ≤9 amino acids) | 101 (17.47%) | 13 (1.85%) |
| ≥60 (epitopes have >9 and ≤15 amino acids) | |||
| >50 (epitopes have >15 amino acids) | |||
Different PID cut-off was used to search epitopes in proteins, cut-off was also set based on amino acids in IgE epitopes.
Motif-based prediction (MEME/MAST)
First, five MEME matrices have been created corresponding to five sets, one matrix for each set. Then each matrix was used as an input file for searching motifs in the remaining four sets using the program MAST. Finally we computed the performance of this approach and achieved a sensitivity in the range of 7% (at 0.001 E-value) to 94% (at 100 E-value) (Table 4). Although the sensitivity increased with increase in E-value, the percent of wrong assignment of non-allergens to allergens also increased from 2.85 to 66.86%. This demonstrates that the motif-based approach developed in this study has a low specificity.
Table 4.
MEME/MAST results of allergen and non-allergen motifs
| E-value | Total hits | |
|---|---|---|
| Allergen | Non-allergen | |
| 10−3 | 38 (6.57%) | 20 (2.86%) |
| 10−1 | 86 (14.88%) | 62 (8.86%) |
| 1 | 142 (24.57%) | 113 (16.14%) |
| 10 | 246 (42.56%) | 240 (34.29%) |
| 20 | 309 (53.46%) | 288 (41.14%) |
| 50 | 427 (73.88%) | 389 (55.57%) |
| 100 | 543 (93.94%) | 468 (66.86%) |
It shows allergen hits out of total 578 allergens and non-allergen hits out of total 700 non-allergens.
Prediction using ARPs
Recently, Bjorklund et al. (12) created a dataset of ARPs, which has 2890 24mers peptides, derived from allergens and non-allergens. The query protein was searched against ARP database using BLAST (36) and assigned allergen if it had similarity with any ARP. In order to avoid any bias, this approach has not been tested on the main dataset, as these allergens were used by Bjorklud et al. (12) for creating ARPs. The approach was tested on 664 allergens and 700 non-allergens (15). As shown in Table 5, the number of correctly assigned allergens (or sensitivity) increased from 52.71 to 94.28% when E-value BLAST increased from 10−9 to 1.0. Though the sensitivity increased with increase in the E-value, the number of non-allergens falsely predicted as allergens also increased from 2 to 338. It was found that the E-value of 0.001 provides a reasonably high sensitivity of 83.58% with only 15 false positive (non-allergens predicted as allergens). Thus the E-value 0.001 was used as a default cut-off for the further studies.
Table 5.
The search results of 1364 proteins (664 allergens and 700 non-allergens), which searched against ARPs database using BLAST
| E-value | Total hits | |
|---|---|---|
| Allergen | Non-allergen | |
| 1 | 626 (94.28%) | 338 (48.25%) |
| 10−1 | 586 (88.22%) | 48 (6.86%) |
| 10−2 | 562 (84.64%) | 23 (3.29%) |
| 10−3 | 555 (83.58%) | 15 (2.14%) |
| 10−4 | 527 (79.37%) | 8 (1.14%) |
| 10−6 | 465 (70.03%) | 5 (0.71%) |
| 10−9 | 350 (52.71%) | 2 (0.28%) |
The boldface indicates the best result
Hybrid approach
The objective of this approach is to improve the sensitivity as well as the specificity of the allergen prediction method. Each approach has its own limitations, as some provide high sensitivity but low specificity and vice verse. In order to get a high sensitivity without loosing much specificity or high specificity with reasonable percent coverage, we combined two or more approaches. First, SVM and IgE epitope-based approaches were combined, and a protein was assigned as an allergen if it was predicted to be one by the IgE method (PID865) and also had a SVM score ≥−0.5. A protein was considered an allergen or a non-allergen using SVM approach, if it had no similarity with any known IgE epitopes. As shown in Table 6, the sensitivity increased around 11% (33.74–44.52), where as the specificity decreased marginally by 0.15%. This shows that one might achieve better sensitivity without loosing much specificity (Supplementary Figures S1 and S2). Similar trend was observed when SVM based method, using dipeptide composition, was combined with the IgE epitope-based method (Supplementary Table S2). No improvement was observed when the motif-based and SVM-based approaches were combined (Supplementary Table S3).
Table 6.
The performance of hybrid approach, which combines SVM-based approach using amino acid composition and IgE epitope based approach (PID865)
| Threshold | Sensitivity | Specificity | Accuracy | PPV | NPV | MCC |
|---|---|---|---|---|---|---|
| 1.0 | 0.4452 | 0.9814 | 0.7396 | 0.9517 | 0.6836 | 0.5211 |
| 0.8 | 0.4922 | 0.9700 | 0.7545 | 0.9309 | 0.7000 | 0.5405 |
| 0.6 | 0.5652 | 0.9586 | 0.7812 | 0.9181 | 0.7293 | 0.5829 |
| 0.4 | 0.6191 | 0.9386 | 0.7945 | 0.8922 | 0.7509 | 0.5995 |
| 0.2 | 0.6713 | 0.9243 | 0.8102 | 0.8793 | 0.7749 | 0.6248 |
| 0.0 | 0.7443 | 0.8929 | 0.8259 | 0.8509 | 0.8106 | 0.6499 |
| −0.2 | 0.8417 | 0.8543 | 0.8486 | 0.8259 | 0.8692 | 0.6963 |
| −0.4 | 0.8887 | 0.8186 | 0.8502 | 0.8009 | 0.9009 | 0.7053 |
| −0.6 | 0.9061 | 0.7614 | 0.8267 | 0.7573 | 0.9096 | 0.6680 |
| −0.8 | 0.9391 | 0.6957 | 0.8055 | 0.7171 | 0.9347 | 0.6441 |
| −1.0 | 0.9583 | 0.6100 | 0.7671 | 0.6687 | 0.9489 | 0.5933 |
Evaluation on an independent dataset
It has been shown in a number of studies that there is a biasness in performance of the method if it is trained and tested on the same dataset, despite n-fold cross-validation (37,38). Thus it is advisable to test any newly developed method on an independent dataset, one not used in the training or the testing of the method. In order to avoid any biasness we used default parameters for each approach (cut-off and the like). As shown in Table 7, the accuracy of prediction based on SVM based approaches were around 85%, followed by ARPs BLAST of 67% and around 93.50% when all approaches have been combined (Supplementary Table S4).
Table 7.
Performance of different methods on 101725 non-allergens obtained from Swiss-Prot and on 323 allergens (independent dataset not used in training or testing of methods).
| Prediction methods | 101 725 non-allergens obtained from Swiss-Prot | Independent dataset of 323 allergens | |
|---|---|---|---|
| Falsely predicted allergens | Specificity (predicted non-allergens) (%) | Allergens correctly predicted allergens (sensitivity) | |
| SVMc | 44684 | 56.07 | 272 (84.21%) |
| SVMd | 39590 | 61.09 | 274 (84.83%) |
| MAST (ev100) | 13545 | 86.68 | 58 (17.95%) |
| MAST (ev 0.1) | 3480 | 96.58 | 40 (12.38%) |
| BLAST (ARP) | 2060 | 97.97 | 215 (66.56%) |
| IgE epitope | 1777 | 98.25 | 35 (10.84%) |
Performance on Swiss-Prot
The performance of the methods was evaluated on a large number of proteins obtained from Swiss-Prot, in order to understand the limitations of methods developed in this study. The aim of this evaluation was to compute the rate of false positive prediction of different methods. We predicted allergens using different approaches at the default threshold in 101 725 non-allergens obtained from Swiss-Prot database (Materials and Methods). The SVM-based method using amino acid and dipeptide composition, falsely predicted 46.74 and 39.30% non-allergens as allergens, respectively. Though specificity of this SVM-based method was poor, the coverage and the sensitivity were higher than those of other methods. In reverse, IgE epitope and MEME methods predicted low rate of false positive but had poor sensitivity.
Comparison with existing methods
Recently, Bjorklund et al. (12), Li et al. (15) and Stadler and Stadler (17) have studied the performance of existing prediction methods and observed that its difficult to compare these methods as different dataset and criteria were used. The aim of this work is to develop complementary method to existing methods for predicting allergenic proteins with high recall and precession. Following are the major advantages of the method (AlgPred server) developed in this study; (i) a large and highly annotated dataset of allergens and non-allergens was used for its development; (ii) it combines existing state-of-art motif based techniques (e.g. MIME/MAST and ARPs); (iii) machine learning technique (SVM) has been introduced to predict allergens with high sensitivity and specificity; (iv) a novel strategy is introduced to predict allergens based on the presence of an IgE epitope, which allows to predict ∼17% of the allergens with low rate of false prediction and (v) different strategies were combined to improve the prediction with high accuracy. Although several methods have been described in the literature, only a limited number of them are available to the public (19,39). Therefore we developed a web server AlgPred, which implements all approaches described in this study. This server will be very useful for biologists especially, in the field of allergy.
Description of the web server
A web server AlgPred has been developed that allows prediction of allergens from amino acid sequence of the protein. The server uses the readseq program (http://iubio.bio.indiana.edu/soft/molbio/readseq/) and accepts the protein sequences in any standard format like EMBL, GCG and FASTA or as plain text format. It allows users to choose any of the following approaches; (i) scanning of IgE epitopes; (ii) motif-based approach; (iii) SVM-based method using amino acid composition of protein; (iv) Hybrid approach; and (v) BLAST search on ARPs. Users can select one or more approaches at a time in a submission form, which allows user to present results of various approaches in a single output. It provides comprehensive information about the prediction that includes score, threshold, distance from threshold, PPV and NPV. If the PPV is >80%, then there is a high chance that the protein is a potential allergen. In case of BLAST search, if the query sequence matches with any ARP in the database, then the matched ARP is also shown. A snapshot of the home page and the sequence submission page of the server is shown in Figure 2a and b. AlgPred also allows the mapping of IgE epitopes on allergenic proteins. The output of AlgPred has been shown in Figure 2c, which contains mapping of IgE epitope, output SVM-based approach and BLAST search against ARPs. The server and related information is available from www.imtech.res.in/raghava/algpred/.
Figure 2.
(a) Snapshot of home page of AlgPred server (b) Snapshot of input page of AlgPred server. (c) Snapshot of output results.
DISCUSSIONS
The aim of this study was to try different approaches for predicting allergenic proteins, particularly food allergens. It agrees with the guidelines of FAO/WHO 2003, according to which the use of multiple tests rather than on a single test is recommended for predicting the allergenic property of the proteins (5,9,10). Here, we tried old as well as new approaches in order to understand their strength and weaknesses. The machine learning technique SVM was used for the first time in this study for predicting allergens. The SVM-based method has been developed using amino acid and dipeptide composition. The major advantage of this approach is that it allows the prediction of allergenic proteins with high sensitivity or specificity, where the user can select cut-off threshold based on his requirement. It was observed that the performance of the dipeptides-based method was lower than that of the amino acid composition-based method, though dipeptides provide more information than amino acid composition. We examined our results and found that most of the allergens used in this study are small so the frequency of occurrence of most of dipeptides was very low or zero.
In the past a number of studies have suggested that identification of epitopes (B or T cell), particularly IgE epitopes, in proteins will be useful in the prediction of allergencity of a protein (23,40–43). Thus an attempt was made in this study to predict allergenic proteins based on the presence of IgE epitopes. Though the aim was to develop accurate method for predicting IgE epitopes, so that allergenic proteins can be predicted, unfortunately, we failed to develop accurate prediction method for IgE epitopes. Thus we used sequence similarity approach, where a protein is assigned allergen if it has high sequence similarity with any known IgE. Though this approach provided high specificity, it had a poor sensitivity compared with the SVM-based approach. This IgE epitope-based strategy helped in improving the performance of the SVM-based method by increasing the sensitivity without reducing the specificity significantly. Ivanciuc et al. (44) developed a method for predicting allergens based on IgE epitopes, in which a sequence is tested by comparing it with IgE epitopes, profiles of IgE epitopes and by surface similarity. It is difficult to compare their method with our method, as the datasets used in the two studies are different. Stadler and Stadler (17) used for the first time a motif-based approach for predicting allergens applying the MEME/MAST algorithm. We applied the same motif-based approach but our results are different from the results reported by Stadler and Stadler (17). This may be due to a number of reasons; (i) the datasets used in the two studies are different; (ii) Stadler and Stadler (17) used generalized profile, where as we used simple MEME/MAST.
The ARPs-based strategy, used in this study to predict allergens, is different from the original method (15). We used BLAST for similarity searching because it is fast and reliable. As shown in the result section, this approach predicts allergens with high accuracy. The web server developed, based on this study, will be very useful for researchers working in the field of food allergens. This server allows the user to predict allergens using any one of the proposed approaches or a combined approach. It is a well-known fact, supported by this study and elsewhere in the literature, that every method has its own limitations. Thus users are advised to use a number of approaches instead of a single approach for predicting allergencity of a protein. A protein predicted to be an allergen by most of the approaches (consensus prediction) has a high probability that it is an allergen.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
Acknowledgments
Authors are thankful to Drs Anastas Pashov (UAMS Little Rock, AR) and Karthikeyan Subramanian (IMTECH Chandigarh, India) for critically reading the manuscript. We are grateful to authors of Bjorklund et al. for providing dataset of allergens and non-allergens as well as ARPs. We are also grateful to authors of Li et al. for providing dataset of 664 allergens used in their study. We acknowledge the financial support from the Council of Scientific and Industrial Research (CSIR) under CMM-017. Funding to pay the Open Access publication charges for this article was provided by CSIR.
Conflict of interest statement. None declared.
REFERENCES
- 1.Sadok Y. Diagnosis of type I hypersensitivity by laboratory test. Tunis Med. 2005;83:441–444. [PubMed] [Google Scholar]
- 2.Scheurer S., Son D.Y., Boehm M., Karamloo F., Franke S., Hoffmann A., Haustein D., Vieths S. Cross-reactivity and epitope analysis of Pru a 1, the major cherry allergen. Mol. Immunol. 1999;36:155–167. doi: 10.1016/s0161-5890(99)00033-4. [DOI] [PubMed] [Google Scholar]
- 3.Santos A.B., Chapman M.D., Aalberse R.C., Vailes L.D., Ferriani V.P., Oliver C., Rizzo M.C., Naspitz C.K., Arruda L.K. Cockroach allergens and asthma in Brazil: identification of tropomyosin as a major allergen with potential cross-reactivity with mite and shrimp allergens. J. Allergy Clin. Immunol. 1999;104:329–337. doi: 10.1016/s0091-6749(99)70375-1. [DOI] [PubMed] [Google Scholar]
- 4.Broadfield E., McKeever T.M., Scrivener S., Venn A., Lewis S.A., Britton J. Increase in the prevalence of allergen skin sensitization in successive birth cohorts. J. Allergy Clin. Immunol. 2002;109:969–974. doi: 10.1067/mai.2002.124772. [DOI] [PubMed] [Google Scholar]
- 5.Goodman R.E., Hefle S.L., Taylor S.L., Ree R.V. Assessing genetically modified crops to minimize the risk of increased food allergy: a review. Int. Arch. Allergy Immunol. 2005;137:153–166. doi: 10.1159/000086314. [DOI] [PubMed] [Google Scholar]
- 6.Taylor S.L. Protein allergenicity assessment of foods produced through agricultural biotechnology. Annu. Rev. Pharmacol. Toxicol. 2002;42:99–112. doi: 10.1146/annurev.pharmtox.42.082401.130208. [DOI] [PubMed] [Google Scholar]
- 7.Lee Y.H., Sinko P.J. Oral delivery of salmon calcitonin. Adv Drug Deliv Rev. 2000;42:225–238. doi: 10.1016/s0169-409x(00)00063-6. [DOI] [PubMed] [Google Scholar]
- 8.Soltero R., Ekwuribe N. The oral delivery of protein and peptide drugs. Innovat. Pharmaceut. Technol. 2002;1:106–110. [Google Scholar]
- 9.FAO/WHO. Evaluation of allergenicity of genetically modified foods. 2001. Report of a joint FAO/WHO expert consultation on allergenicity of foods derived from biotechnology ( http://www.fao.org/es/ESN/food/pdf/allergygm.pdf)
- 10.FAO/WHO. Report of the fourth session of the codex ad hoc intergovernmental task force on foods derived from biotechnology. 2003. ( http://www.codexalimentarius.net/download/report/46/Al0334ae.pdf) [PubMed]
- 11.Hileman R.E., Silvanovich A., Goodman R.E., Rice E.A., Holleschak G., Astwood J.D., Hefle S.L. Bioinformatic methods for allergenicity assessment using a comprehensive allergen database. Int. Arch. Allergy Immunol. 2002;128:280–291. doi: 10.1159/000063861. [DOI] [PubMed] [Google Scholar]
- 12.Bjorklund A.K., Soeria-Atmadja D., Zorzet A., Hammerling U., Gustafsson M.G. Supervised identification of allergen-representative peptides for in silico detection of potentially allergenic proteins. Bioinformatics. 2005;21:39–50. doi: 10.1093/bioinformatics/bth477. [DOI] [PubMed] [Google Scholar]
- 13.Gendel S.M. Sequence analysis for assessing potential allergenicity. Ann. NY Acad. Sci. 2002;964:87–98. doi: 10.1111/j.1749-6632.2002.tb04134.x. [DOI] [PubMed] [Google Scholar]
- 14.Kleter G.A., Peijnenburg A.A. Screening of transgenic proteins expressed in transgenic food crops for the presence of short amino acid sequences identical to potential, IgE-binding linear epitopes of allergens. BMC Struct. Biol. 2002;2:8. doi: 10.1186/1472-6807-2-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li K.B., Issac P., Krishnan A. Predicting allergenic proteins using wavelet transform. Bioinformatics. 2004;20:2572–2578. doi: 10.1093/bioinformatics/bth286. [DOI] [PubMed] [Google Scholar]
- 16.Silvanovich A., Nemeth M.A., Song P., Herman R., Tagliani L., Bannon G.A. The value of short amino acid sequence matches for prediction of protein allergenicity. Toxicol. Sci. 2006;90:252–258. doi: 10.1093/toxsci/kfj068. [DOI] [PubMed] [Google Scholar]
- 17.Stadler M.B., Stadler B.M. Allergenicity prediction by protein sequence. FASEB J. 2003;17:1141–1143. doi: 10.1096/fj.02-1052fje. [DOI] [PubMed] [Google Scholar]
- 18.Aalberse R.C. Structural biology of allergens. J. Allergy Clin. Immunol. 2000;106:228–238. doi: 10.1067/mai.2000.108434. [DOI] [PubMed] [Google Scholar]
- 19.Fiers M.W., Kleter G.A., Nijland H., Peijnenburg A.A., Nap J.P., Van H.R.C. Allermatch, a webtool for the prediction of potential allergenicity according to current FAO/WHO Codex alimentarius guidelines. BMC Bioinformatics. 2004;5:133. doi: 10.1186/1471-2105-5-133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zorzet A., Gustafsson M., Hammerling U. Prediction of food protein allergenicity: a bioinformatic learning systems approach. In Silico Biol. 2002;2:525–534. [PubMed] [Google Scholar]
- 21.Soeria-Atmadja D., Zorzet A., Gustafsson M.G., Hammerling U. Statistical evaluation of local alignment features predicting allergenicity using supervised classification algorithms. Int. Arch. Allergy Immunol. 2004;133:101–112. doi: 10.1159/000076382. [DOI] [PubMed] [Google Scholar]
- 22.Ivanciuc O., Schein C.H., Braun W. SDAP: database and computational tools for allergenic proteins. Nucleic Acids Res. 2003;31:359–362. doi: 10.1093/nar/gkg010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Saha S., Raghava G.P.S. BcePred: prediction of continuous B-cell epitopes in antigenic sequences using physico-chemical properties. In: Nicosia G., Cutello V., Bentley P.J., Timis J., editors. Artificial Immune Systems, Third International Conference (ICARIS 2004), LNCS 3239. Italy: Catania, Sicily; 2004. pp. 197–204. [Google Scholar]
- 24.Blythe M.J., Flower D.R. Benchmarking B cell epitope prediction: underperformance of existing methods. Protein Sci. 2005;14:246–248. doi: 10.1110/ps.041059505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Saha S., Bhasin M., Raghava G.P.S. Bcipep: a database of B-cell epitopes. BMC Genomics. 2005;6:79. doi: 10.1186/1471-2164-6-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bendtsen J.D., Jensen L.J., Blom N., Von H.G., Brunak S. Feature-based prediction of non-classical and leaderless protein secretion. Protein Eng. Des. Sel. 2004;17:349–356. doi: 10.1093/protein/gzh037. [DOI] [PubMed] [Google Scholar]
- 27.Brendel V. PROSET—a fast procedure to create non-redundant sets of protein sequences. Math. Comput. Model. 1992;16:37–43. [Google Scholar]
- 28.Boeckmann B., Bairoch A., Apweiler R., Blatter M.C., Estreicher A., Gasteiger E., Martin M.J., Michoud K., O'Donovan C., Phan I., et al. The Swiss-Prot protein knowledgebase and its supplement TrEMBL in 2003. Nucleic Acids Res. 2003;31:365–370. doi: 10.1093/nar/gkg095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Timothy L.B., Charles Elkan. ‘Fitting a mixture model by expectation maximization to discover motifs in biopolymers’. Proceedings of the Second International Conference on Intelligent Systems for Molecular Biology; Menlo Park, CA: AAAI Press; 1994. pp. 28–36. [PubMed] [Google Scholar]
- 30.Timothy L.B., Michael Gribskov. ‘Combining evidence using P-values: application to sequence homology searches’. Bioinformatics. 1998;14:48–54. doi: 10.1093/bioinformatics/14.1.48. [DOI] [PubMed] [Google Scholar]
- 31.Joachims T. Making large-scale SVM learning particle. In: Scholkopf B., Burges C., Smola A., editors. Advances in Kernal Methods Support Vector Learning. Cambridge, MA and London: MIT Press; 1999. pp. 42–56. [Google Scholar]
- 32.Baldi P., Brunak S., Chauvin Y., Andersen C.A., Nielsen H. Assessing the accuracy of prediction algorithms for classification: an overview. Bioinformatics. 2000;16:412–424. doi: 10.1093/bioinformatics/16.5.412. [DOI] [PubMed] [Google Scholar]
- 33.Bhasin M., Raghava G.P.S. Classification of nuclear receptors based on amino acid composition and dipeptide composition. J. Biol. Chem. 2004;279:23262–232626. doi: 10.1074/jbc.M401932200. [DOI] [PubMed] [Google Scholar]
- 34.Garg A., Bhasin M., Raghava G.P.S. Support vector machine-based method for subcellular localization of human proteins using amino acid compositions, their order, and similarity search. J. Biol. Chem. 2005;280:14427–14432. doi: 10.1074/jbc.M411789200. [DOI] [PubMed] [Google Scholar]
- 35.Kumar M., Verma R., Raghava G.P.S. Prediction of mitochondrial proteins using support vector machine and hidden markov model. J. Biol. Chem. 2005;281:5357–5363. doi: 10.1074/jbc.M511061200. [DOI] [PubMed] [Google Scholar]
- 36.Altschul S.F., Madden T.L., Schaffer A.A., Zhang J., Zhang Z., Miller W., Lipman D.J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kumar M., Bhasin M., Natt N.K., Raghava G.P.S. BhairPred: prediction of beta-hairpins in a protein from multiple alignment information using ANN and SVM techniques. Nucleic Acids Res. 2005;33:W154–W159. doi: 10.1093/nar/gki588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Soeria-Atmadja D., Wallman M., Bjorklund A.K., Isaksson A., Hammerling U., Gustafsson M.G. External cross-validation for unbiased evaluation of protein family detectors: application to allergens. Proteins. 2005;61:918–925. doi: 10.1002/prot.20656. [DOI] [PubMed] [Google Scholar]
- 39.Riaz T., Hor H.L., Krishnan A., Tang F., Li K.B. WebAllergen: a web server for predicting allergenic proteins. Bioinformatics. 2005;21:2570–2571. doi: 10.1093/bioinformatics/bti356. [DOI] [PubMed] [Google Scholar]
- 40.Brusic V., Petrovsky N., Gendel S.M., Millot M., Gigonzac O., Stelman S.J. Computational tools for the study of allergens. Allergy. 2003;58:1083–1092. doi: 10.1034/j.1398-9995.2003.00224.x. [DOI] [PubMed] [Google Scholar]
- 41.Bhasin M., Singh H., Raghava G.P.S. MHCBN: a comprehensive database of MHC binding and non-binding peptides. Bioinformatics. 2003;19:665–666. doi: 10.1093/bioinformatics/btg055. [DOI] [PubMed] [Google Scholar]
- 42.Singh H., Raghava G.P.S. ProPred: prediction of HLA-DR binding sites. Bioinformatics. 2001;17:1236–1237. doi: 10.1093/bioinformatics/17.12.1236. [DOI] [PubMed] [Google Scholar]
- 43.Singh H., Raghava G.P. ProPred1: prediction of promiscuous MHC Class-I binding sites. Bioinformatics. 2003;19:1009–1014. doi: 10.1093/bioinformatics/btg108. [DOI] [PubMed] [Google Scholar]
- 44.Ivanciuc O., Mathura V., Midoro-Horiuti T., Braun W., Goldblum R.M., Schein C.H. Detecting potential IgE-reactive sites on food proteins using a sequence and structure database, SDAP-food. J. Agric Food Chem. 2003;51:4830–4837. doi: 10.1021/jf034218r. [DOI] [PubMed] [Google Scholar]