Abstract
PHEPS (pH-dependent Protein Electrostatics Server) is a web service for fast prediction and experiment planning support, as well as for correlation and analysis of experimentally obtained results, reflecting charge-dependent phenomena in globular proteins. Its implementation is based on long-term experience (PHEI package) and the need to explain measured physicochemical characteristics at the level of protein atomic structure. The approach is semi-empirical and based on a mean field scheme for description and evaluation of global and local pH-dependent electrostatic properties: protein proton binding; ionic sites proton population; free energy electrostatic term; ionic groups proton affinities (pKa,i) and their Coulomb interaction with whole charge multipole; electrostatic potential of whole molecule at fixed pH and pH-dependent local electrostatic potentials at user-defined set of points. The speed of calculation is based on fast determination of distance-dependent pair charge-charge interactions as empirical three exponential function that covers charge–charge, charge–dipole and dipole–dipole contributions. After atomic coordinates input, all standard parameters are used as defaults to facilitate non-experienced users. Special attention was given to interactive addition of non-polypeptide charges, extra ionizable groups with intrinsic pKas or fixed ions. The output information is given as plain-text, readable by ‘RasMol’, ‘Origin’ and the like. The PHEPS server is accessible at http://pheps.orgchm.bas.bg/home.html.
INTRODUCTION
Electrostatic phenomena are widely manifested as a fundamental feature of protein structure–function relationships (1–5). Protein molecules are very complex dielectric systems but can be treated as ‘solid state’ nanometer particles, immersed in buffered water solutions. Many approaches [‘macroscopic’—continuum dielectrics (6,7) and ‘microscopic’—polarizability (8,10)] were developed with different degree of validity: from simple TK-‘dielectric cavity models’ and analytical solution of Poisson–Boltzmann equation to their non-linear numerical and sophisticated empirical generalized Born solutions (11–13). The application of detailed and complex model description leads to increased difficulties for experimentalists to understand and use such sophisticated models. At present there are number of popular program packages [(14,15) and others] and web servers (16,17) but they are of limited significance for everyday problems of experimentalists. To the best of our knowledge, there is not available web server for fast pH-dependent calculation and analysis of protein electrostatic properties. Such software is needed because proteins are polyelectrolytes and their system of ionizable groups is pH-dependent. Many programs and servers compute pKas at ‘neutral pH’ yielding pH-independent pKas, which leads to erroneous results and distorts our view of principal properties of protein molecules, important for their functions. It is known that ‘pKa’ is directly related to free energy change of the corresponding protolytic reaction (ΔGa = RTpKa) and that this ΔGa is pH-dependent. It is well known that protein pKas are also pH-dependent, because ionic groups are closely arranged in the molecule. There are excellent theoretical works describing pH-dependent protein electrostatics [(7,18,19) and so on] but they are not straightforward for application by experimentalists. For many years a method addressing aforementioned requirements was developed and applied successfully in Biophysical Chemistry Laboratory at IOCh. The theoretical results are unequivocally validated by comparison with experimental studies as shown in a number of peer-reviewed publications over the years. Typical examples for this are pKas prediction of lysozyme, BPTI and cytochrome c (21–23); spectrophometric titration prediction and infrared carboxylic groups titration (24), enthalpy of protein ionization prediction (25), pH-dependent protein ultrasonic compressibility analysis (26); local electrostatic potentials (27); electrostatic contribution to a protein crystal lattice energy (28) and so on. The method was also applied to clarify enzyme mechanisms (29) and proved to be an invaluable tool for fast evaluation of electrostatic interactions and their analysis in large biomolecular immunochemical complexes (30). We hope that our server http://pheps.orgchm.bas.bg/home.html will be useful for experimentalist (protein scientist in need for fast evaluation of pH-dependent properties, enzymologists in need of pK values, spectroscopists and the like) as well as for in silico analysis by structural biologists and bioinformaticians. Being fast and easy to use this server is suitable for first acquaintance and training in the field.
METHODS
Protein self-consistent electrostatics
It is generally accepted that a model for protein electrostatics can be build on the assumption of continuum medium description, fixed atom approximation, protein–solvent boundary numerically described by atomic static accessibilities, SAi [variants of Lee-Richards algorithm (31)] and two type of charges: (i) permanent (pH-independent) partial charges (par) and (ii) proton-binding sites with pH-dependent titratable charges (tit). The model accepts experimentally measured pKa of model compounds (e.g. N-acetyl amides of each i-th ionogenic amino acids) (pKmod,i) and evaluates work for charge transfer from highly polar water solvent (ɛw = 80) to protein macromolecule (4 < ɛp,i < 40). Exposure to the solvent is evaluated by SAi at absence of other ionic groups. Born term, which is proportional to [1 − (ɛp,i/ɛw)], is roughly estimated as (1 − SAi). Partial charges assume values from AMBER and PARSE parameterization sets. Since the ratio of the number of ionic AAR (Nion) to total number of AAR (Ntot) Rel = Nion/Ntot is relative high for protein particles with small radii (Rp), the pairwise interaction between any i-th and j-th ionic groups counts contributions from charge–charge, charge–dipole and dipole–dipole interactions which can be simulated by an empirical three exponential curve:
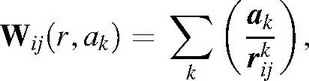 |
where k = 1 for long-range (Columbic) interactions; k = 2 for mid-range, charge–dipole interactions; and k = 3 for short-range dipole–dipole interactions. The ak were estimated by a non-linear procedure by minimizing the functional F(a1,a2,a3) (21):
where the values of Zexp are taken from experimental data and Zth are the calculated values of the protein net charge as a function of pH. The initial values of the coefficients ak are obtained by numerical approximation of W(rij). Through extensive testing, using large dataset of structures, it was found that a1, a2 and a3 values are practically constants for a great number of proteins.
The pH-dependence of the electrostatic potential Φel,i (pH) at the i-th proton binding site in PHEI was evaluated according to the following equation:
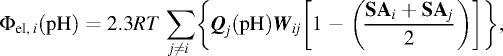 |
where Qj(pH) is defined by degree of dissociation or statistical mechanical proton population of given H+-binding site; Qj (pH) = (1 − 〈sj〉) and Qj (pH) = −〈sj〉 for basic and acidic groups respectively, where
Thus using partial titration of each j-th group we can find the pH-dependent net-charge of the whole molecule, Z(pH), i.e. potentiometric titration curve:
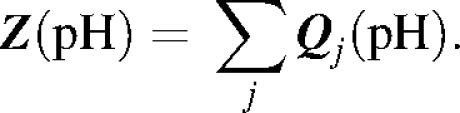 |
By definition if Z = 0 than pH = pI, i.e. the isoelectric point (the only pH at which the dipole moment of a protein molecule can be evaluated).
Thus starting with pKint,i = pKmod,i + ΔpKBorn,i + ΔpKpar,i, where pKmod,i is the pKa of the i-th site according to model compounds—see set of pKmod,i in (21,22,29); ΔpKBorn,i is the Born self-energy of the i-th site buried within the ‘uncharged’ protein, and ΔpKpar,i is the contribution of the i-th site interacting with the set of partial (permanent, fixed) atomic charges (see above).
where C is the Debye–Hückel term for ionic strength (Is). The term pKtit,i is the pKa shift of the i-th site caused by interactions with all other proton-binding groups and is evaluated according to efficient self-consistent iterative procedure (32). Coming to self consisted pH-dependent ionization the free energy term Gel(pH) is calculated as follows.
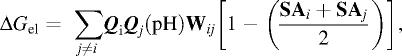 |
as well as pH-dependent Coulomb energy of each i-th ionic group with whole charge multipole:
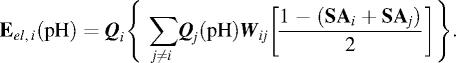 |
After applying this iterative algorithm the electrostatic system is converged and all basic pH-dependent properties are reported.
IMPLEMENTATION
The web sever is a front end of our program package PHEI, developed over many years in our Biophysical Chemistry Lab. Its current version is written in PERL and C/C++ by one of us (A.K.). Our package is capable of much more functionality and only basic electrostatic properties are presented online, the rest being under consideration for the next release (Conclusions and Future). The web implementation is driven by CGI/PERL routines. The only input file is a coordinate file in Protein Data Bank (PDB) format (33)—either user supplied or just as a PDB ID, following retrieval from our local PDB database. Following submission, the user is given some basic information about the protein molecule (chains; number of residues; ratio of ionogenic to all groups, Rel) and warned about certain inconsistencies in structure, related to subsequent calculation (interruption in residue numbering which might influence appearance of terminal charges). The user is given the possibility to edit initial setup of ionogenic groups (attention to CYS in SSBONDs and excluding covalently modified groups). This is accomplished by convenient interactive selection of used set of groups. This gives opportunity for simulation of ‘electrostatic mutagenesis’. Full ‘charge mutant analysis’ is supposed for next versions. The same screen visualizes the PDB file in a text field which allows for direct editing: adding missing terminal charges, fixed (non-titratable) whole or partial charges and titratable groups with user defined pKa intrinsic. All other parameters used as input are predefined or automatically calculated. After initial setup completion the calculation proceeds through aforementioned steps—evaluation of accessibilities and Born term ΔpKBorn,i, perturbation of pKa by partial charges ΔpKpar,i and finally the iterative procedure for self-consistent evaluation of titratable ΔpKtit,i.
To calculate ΔGel(pH), the following energy conversion units were used: 1 kcal = 4.186 kJ = 1.68RT units (at 298 K) = 0.735 pKa units. The units of ϕi(pH) are kcal/mol·e = 43.176 mV or 30.24 mC/m2. All calculations are provided at ionic strength (Is) 0.1.
The obtained results are organized in two groups: (i) GLOBAL [Z(pH), ΔGel(pH) and Φj at fixed pH] and (ii) LOCAL [si(pH), pKa,i(pH), Eel,i(pH) and ϕi(pH)]. For each of them there is a link to own page. The contents of each page is comprised of the result itself, related derivatives (e.g. pI, ∂Z/∂pH, pK1/2 and so on) as well as a short description and examples for visualization of this type of data. All output data files are in standard plain text format. Visualization is straightforward with any 2D plotting software and molecular graphics programs (RasMol, JMol, PyMol and so on).
RESULTS
Global pH-characteristics
(1) pH-dependent protein net charge Z(pH) and its derivatives: Isoelectric point pI/Z = 0 and protein buffer capacity β = ∂pI/∂pH at three pH: (pI − 1.5), pI and (pI + 1.5).
It is equivalent to experimental potentiometric titration curve (34) and reflects basic global electrostatic characteristic of protein proton binding (35). The definition of pI is pH at which Z = 0. Protein buffer capacity (β) is an important parameter for design of precise ion-exchange (36) and electrophoresis (37) experiments. The difference between two Z(pH) of analogous but perturbed states [e.g. apo-holo (30), oxidized-reduced, free-liganded and the like] can be useful in analysis the nature of such perturbation and identify pH region where it has maximal effect on proton binding. Other relevant issues are: the net charge of protein under condition of electro-spray mass-spectrometry (38); the critical Z-values at extreme pH in water (39,40) and in vacuum (41) at which protein ‘denature’ and many others (Supplementary Figure S1).
(2) pH-dependent electrostatic free energy term [ΔGel(pH)] and its derivatives: ΔGel,min, ΔGel,pI; pHa and pHb at ΔGel = 0 for acid and alkaline/basic denaturation, respectively.
Quantitative estimate for charge dependent stability ΔGel(pH) is basic electrostatic characteristic of protein molecules (2). By evaluating ΔΔGel,ion(pH) = ΔGel,holo(pH) − ΔGel,apo(pH) it is possible to determine pH-dependent specific ion and/or cofactor binding (30). Similarly ‘electron affinity’ can be evaluated from difference ΔΔGel,e(pH) = ΔGel,red(pH) − ΔGel,oxid(pH) (42) and the like. It is easy to obtain experimental values for pHd,a and pHd,b and compare with calculated by our server (43). Another option is estimation of stability of pH-induced conformational states and evaluation of energetic barrier between them (44). Presence or absence of stricture ruled charge asymmetry is reflected in ΔGel,min − ΔGel,pI difference (also from their pHmin − pI) (Supplementary Figure S2).
(3) Electrostatic potential, EP(Φel) at user selected pH for all j-th protein non-hydrogen atoms in a PDB-formatted file and can be visualized in color scale by RasMol.
The electrostatic potential at each point within (45), on the molecular surface (46) and at near vicinity in solvent (47) for a protein molecule is its fundamental electrostatic characteristic (8,48). In fact all above quantities are derivative of Φel = f(pH, ligands). Using present PHEPS version output file, it is straightforward to visualize Φel (or EP) at each protein non-hydrogen atoms by switching on ‘color by temperature’ using color scale (dark blue: positive EP; green: zeroed EP; and red: negative EP) applicable to entire variety of RasMol model representations (Supplementary Figure S3).
Local pH-characteristics
(4) pH-dependent proton population or degree of ionization of each i-th ionic group (Si).
The results Si(pH) for ionic groups in order of increasing sequence numbers are presented in the form of column formatted file (all in one table).
Si(pH) can be related to NMR pH-dependent chemical shifts, δi(pH) (49,50) or other individual titration characteristics—FTIR carboxylate titration (51); differential Tyr UV-titration (52); calorimetric/enthalpy titration [ΔHi(pH)] (53) and so on (Supplementary Figure S4).
(5) pH-dependent proton affinity pKa,i(pH) at each individual i-th ionic site: The results pKa,i(pH) for ionic groups in order of increasing sequence numbers are presented in the form of column formatted file (all in one table). The set of pK1/2,i for each i-th ionizable group [their pKas at si(pH) = 0.5] is available in another table.
Predicted pK1/2,i can be compared directly to experimentally obtained. Plotting pKa,i(pH) is a fast way to differentiate ‘normal’ and ‘abnormal’ titration groups (19,53); functionally important ionogenic sites (54); ΔpKda = (pKdonor − pKacceptor) as function of pH to necessary for description pf H+-transfer processes (Hydrogen Bonded Networks, Brønsted's relations and the like) (55,56) (Supplementary Figure S5).
(6) pH-dependent electrostatic energy Eel,i(pH) of interaction of each i-th ionic group with whole multipole of partial and protonic/ionic charges—individual sites and their sum. The results Eel,i(pH) for ionic groups in order of increasing sequence numbers are presented in the form of column formatted file (all in one table).
The pH-dependent Columbic energy of interaction of given ionic group with whole charge multipole is evident characteristic reflecting specific electrostatic site property: influence on charge stability (57), participation in charge-driven processes (58), through space interactions with introduced charged systems in protein complexes (30) and the like (Supplementary Figure S6).
(7) pH-dependent local electrostatic potential, ϕi(pH) at each i-th point within protein molecule or its close surrounding. The user is supposed to define points in PDB format. It is recommended that number of points do not exceed 20. The results are presented as pH-dependence of electrostatic potential at each point.
Knowledge of local EP at user-defined points is a great tool for elucidating electrostatic response of these sites to intra/inter-molecular interactions with charged groups (ions and dipoles of different kind of ligands in static and dynamic manner) (59,60). This characteristic is of indispensable use for evaluation of the effect of whole protein electrostatic field on crucially important sites (e.g. for understanding its role in intermediate species of enzyme catalytic cycles (59), protein stability (60,62) and many others (Supplementary Figure S7).
Tested proteins. All these features of our program package PHEI were developed and have been tested for many years. The method was applied to numerous proteins [Supplementary Table 1, (20–30)]. Calculated pK1/2-s was compared with experimental estimates of pKas (21,22) and correlation was made of calculated Z(pH) to published experimental curves.
CONCLUSION AND FUTURE DEVELOPMENT
We hope this server will be useful to anyone who needs fast and detailed analysis of pH-dependent properties of a protein with known atomic structure and a tool for protein electrostatic design (61,62). We are ready to share our experience in the field with other protein scientists and are open for discussion.
Features in preparation for next PHEPS version are as follows:
For each AA in sequence order n (backbone, side chain and residue) with respective (B-factor)n, static accessibility 〈SA〉n and φel,n.—now implemented.
3D-contour EP map generation (in static and dynamic regime)—search of saddle and other critical points on multidimensional maps.
Correct determination of dipole (at pI) and electric (at any other pH) moments (μd and μe, respectively) using 3D-EP grid—their scalar and vector values.
Thorough ‘electrostatic mutation’ analysis with ‘Δ = mut − wild’ as function of pH—data for all mentioned above characteristics: ΔZ, ΔSi, ΔpKi, ΔΔGel, ΔEel and ΔΦel.
EP gradients (electrostatic forces, EF) at pH control, located at defined atoms and sites (user selected fragments, domains, subunits).
Many of these features are implemented in our program package PHEI, but their online access will be realized after extensive testing.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
Supplementary Material
Acknowledgments
We thank Profs B. Honig and E. Alexov for kind donation of computers, one of which hosts our server. We thank L. Roumenina for her kind help in text edition and correction. This work is partially supported by grant X-1310 of National Fund ‘Scientific Research’, Sofia, Bulgaria. The Open Access publication charges for this article were waived by Oxford University Press
Conflict of interest statement. None declared.
REFERENCES
- 1.Perutz M.F. Electrostatic effects in proteins. Science. 1978;201:1178–1191. doi: 10.1126/science.694508. [DOI] [PubMed] [Google Scholar]
- 2.Honig B., Nicholls A. Classical electrostatics in biology and chemistry. Science. 1995;268:1144–1149. doi: 10.1126/science.7761829. [DOI] [PubMed] [Google Scholar]
- 3.Warshel A. Electrostatic basis of structure–function correlation in proteins. Acc. Chem. Res. 1981;14:284–290. [Google Scholar]
- 4.Bashford D., Karplus M. pKas of ionization groups in proteins: atomic detail from a continuum electrostatic model. Biochemistry. 1990;29:10219–10225. doi: 10.1021/bi00496a010. [DOI] [PubMed] [Google Scholar]
- 5.Antosiewicz J., McCammon J.A., and Gilson M.K. Prediction of pH-dependent properties of proteins. J. Mol. Biol. 1994;238:415–436. doi: 10.1006/jmbi.1994.1301. [DOI] [PubMed] [Google Scholar]
- 6.Simonson T. Macromolecular electrostatics: continuum models and their grooving pains. Curr. Opin. Struct. Biol. 2001;11:243–253. doi: 10.1016/s0959-440x(00)00197-4. [DOI] [PubMed] [Google Scholar]
- 7.Bashford D. Macroscopic electrostatic models for protonation state in proteins. Front. Biosci. 2004;9:1082–1099. doi: 10.2741/1187. [DOI] [PubMed] [Google Scholar]
- 8.Warshel A., Papazyan A. Electrostatic effects in macromolecules: fundamental concepts and practical modeling. Curr. Opin. Struct. Biol. 1998;8:211–217. doi: 10.1016/s0959-440x(98)80041-9. [DOI] [PubMed] [Google Scholar]
- 9.Braun-Sand S., Warshel A. ‘Electrostatics of proteins: models and principles’. In: Buchner J., Kiefhaber T., editors. Protein Folding Handbook, Part I. New York: Wiley-VCH GmbH & Co.; 2005. [Google Scholar]
- 10.Aquist J. Calculation of absolute binding free energies for charged ligands and effects of long-range electrostatic interactions. J. Comput. Chem. 1996;17:1587–1597. [Google Scholar]
- 11.Matthews J.B., Gurd F.R.N. Calculation of electrostatic interactions in proteins. Methods Enzymol. 1986;130:413–453. doi: 10.1016/0076-6879(86)30019-3. [DOI] [PubMed] [Google Scholar]
- 12.Petersen M.T.N., Petersen S.B. How to lipases and esterases work: the electrostatic contribution. J. Biotechnol. 2001;85:115–147. doi: 10.1016/s0168-1656(00)00360-6. [DOI] [PubMed] [Google Scholar]
- 13.Feig M., Onufriev A., Lee M.S., Im W., Case E.A., Brooks C.L., III Performance comparison of generalized Born and Poisson methods in the calculation of electrostatic solvation energies for protein structures. J. Comput. Chem. 2004;25:265–284. doi: 10.1002/jcc.10378. [DOI] [PubMed] [Google Scholar]
- 14.Nayal M., Hitz B.C., Honig B. GRASS: a server for the graphical representation and analysis of structures. Protein Sci. 1999;8:676–679. doi: 10.1110/ps.8.3.676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dolinsky D.J., Nielson J.E., McCammon A.J., Backer N.A. PDB2PQR: an automated pipeline for the setup of Poisson-Boltzmann electrostatic calculations. Nucleic Acid Res. 2004;32:W665–W667. doi: 10.1093/nar/gkh381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gordon J.C., Meyers J.B., Folta T., Shoja V., Heath L.S., Onufriev A. H++: a server for estimating pKas and adding missing hydrogens to macromolecules. Nucleic Acid Res. 2005;33:W368–W371. doi: 10.1093/nar/gki464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Miteva M.A., Tuffery P., Villoutreix B.O. PCE: web tools to compute protein continuum electrostatics. Nucleic Acid Res. 2005;33:W372–W375. doi: 10.1093/nar/gki365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Georgesku R.E., Alexov E., Gunner M. Combining conformational flexibility and continuum electrostatics for calculating pK(a)s in proteins. Biophys. J. 2002;83:1731–1748. doi: 10.1016/S0006-3495(02)73940-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Spassov V.Z., Bashford D. Electrostatic coupling to pH-titration sites are a source of cooperativity in protein-ligand binding. Protein Sci. 1998;7:2012–2025. doi: 10.1002/pro.5560070918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Atanasov B., Karshikov A. Semi-empirical method for calculation of electrostatic interactions in proteins. Studia Biophysica. 1985;105:11–22. [Google Scholar]
- 21.Spassov V.Z., Karshikov A.D., Atanasov B.P. Electrostatic interactions in proteins: a theoretical analysis of lysozyme ionization. Biochim. Biophys. Acta. 1989;999:1–6. [Google Scholar]
- 22.Karshikov A.D., Engh R., Bode W., Atanasov B.P. Electrostatic interactions in proteins: Calculations of the electrostatic term of free energy and the electrostatic potential field. Eur. Biophys. J. 1989;17:287–297. [Google Scholar]
- 23.Kossekova G.P., Miteva M.A., Atanasov B.P. Characterization of pyridoxal phosphate as an optical label for measuring electrostatic potentials in proteins. J. Photochem. Photobiol. B. 1997;37:74–83. doi: 10.1016/1011-1344(95)07211-x. [DOI] [PubMed] [Google Scholar]
- 24.Miteva M., Alexov E., Atanasov B. Numerical simulation of aldolase tetramer stability. Eur. Biophys J. 1998;28:67–73. [Google Scholar]
- 25.Atanasov B., Miteva M. Prediction and structural analysis of the enthalpy of ionisation of proteins. Thermochim. Acta. 1997;291:141–153. [Google Scholar]
- 26.Miteva M.A., Mishonova E.I., Atanasov B.P. A theoretical model for evaluation of the ultrasonic velocimetric titration of proteins. Comput. Rend. Acad. Bulg. Sci. 1996;49:101–104. [Google Scholar]
- 27.Miteva M.A., Kossekova G.P., Villoutreix B.O., Atanasov B.P. Local electrostatic potentials in pyridoxal phosphate labeled horse heart cytochrome c. J. Photochem. Photobiol. B. 1997;37:74–83. doi: 10.1016/s1011-1344(96)07345-9. [DOI] [PubMed] [Google Scholar]
- 28.Alexov E., Atanasov B. Analysis of electrostatic interactions in ribonuclease A monoclinic crystal. Biochim. Biophys. Acta. 1994;1206:55–62. doi: 10.1016/0167-4838(94)90072-8. [DOI] [PubMed] [Google Scholar]
- 29.Atanasov B., Mustafi D., Makinen M.W. Protonation of the β-lactam nitrogen is the trigger event in the catalytic action of class A β-lactamases. Proc. Natl Acad. Sci. USA. 2000;97:3160–3165. doi: 10.1073/pnas.060027897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Roumenina L.T., Kantardjiev A.A., Atanasov B.P., Waters P., Gadjeva M., Reid K.B.M., Mantovani A., Kishore U., Kojouharova M.S. Role of Ca2+ in the electrostatic stability and the functional activity of the globular domain of the human C1q. Biochemistry. 2005;44:14097–14109. doi: 10.1021/bi051186n. [DOI] [PubMed] [Google Scholar]
- 31.Lee B., Richards F.M. The interpretation of protein structures: estimation of static accessibility. J. Mol. Biol. 1971;55:379–400. doi: 10.1016/0022-2836(71)90324-x. [DOI] [PubMed] [Google Scholar]
- 32.Karshikoff A. A simple algorithm for calculation of multiple site titration curves. Protein Eng. 1995;8:243–248. doi: 10.1093/protein/8.3.243. [DOI] [PubMed] [Google Scholar]
- 33.Westbrook J., Feng Z., Chen L., Yang H., Berman H.M. The Protein Data Bank and structural genomics. Nucleic Acid Res. 2003;31:489–491. doi: 10.1093/nar/gkg068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tanford C. The interpretation of hydrogen ion titration curves of proteins. Adv. Protein Chem. 1962;17:69–165. [Google Scholar]
- 35.Nitta K., Sugai Sh. Potentiometric titration studies on globular proteins. Biopolymers. 1972;11:1893–1901. doi: 10.1002/bip.1972.360110909. [DOI] [PubMed] [Google Scholar]
- 36.Egmond M.R., Antheunisse W.P., van Bemmel C.J., Ravestein P., de Vlieg J., Peters H., Branner S. Engineering surface charges in a subtilisin: the effects on electrophoretic and ion-exchange behavior. Protein Eng. 1994;7:793–800. doi: 10.1093/protein/7.6.793. [DOI] [PubMed] [Google Scholar]
- 37.Palusinski O.A., Su Y., Fife P.C. Numerical technique and computational procedure for isotachophoresis. Electrophoresis. 1990;11:903–907. doi: 10.1002/elps.1150111104. [DOI] [PubMed] [Google Scholar]
- 38.Kaltashov I., Eyles S.J. Studies of biomolecular conformations and conformational dynamics by Mass Spectrometry. Mass Spectrom. Rev. 2002;21:37–71. doi: 10.1002/mas.10017. [DOI] [PubMed] [Google Scholar]
- 39.Oliveberg M., Vuilleumier S., Fersht A.R. Thermodynamic study of the acid denaturation of barnase and its dependence on ionic strength: evidence for residual electrostatic interactions in the acid/thermally denatured state. Biochemistry. 1994;33:8826–8832. doi: 10.1021/bi00195a026. [DOI] [PubMed] [Google Scholar]
- 40.Anderson D., Becktel W.J., Dahlquist F.W. pH-induced denaturation of proteins: A single salt bridge contributes 3–5 kcal/mol to the free energy of folding of T4 lysozyme. Biochemistry. 1990;29:2403–2408. doi: 10.1021/bi00461a025. [DOI] [PubMed] [Google Scholar]
- 41.Valentine S.J., Counterman A.E., Clemmer D.E. Conformer dependent proton transfer reactions of Ubiquitin ions. J. Am. Soc. Mass Spectrom. 1997;8:954–961. [Google Scholar]
- 42.Salgueiro C.A., daCosta P.N., Turner D.L., Messias A.C., van Dongen W.M., Saraiva L.M., Xavier A.V. Effect of hydrogen-bond networks in controlling reduction potentials in Desulfovibrio vulgaris (Hildenborough) cytochrome C3 probed by site-specific mutagenesis. Biochemistry. 2001;40:9709–9716. doi: 10.1021/bi010330b. [DOI] [PubMed] [Google Scholar]
- 43.Meiering E.M., Serrano L., Fersht A.R. Effect of active site residues in barnase on activity and stability. J. Mol. Biol. 1992;225:585–589. doi: 10.1016/0022-2836(92)90387-y. [DOI] [PubMed] [Google Scholar]
- 44.Yang A.S., Honig B. Structural origin of pH and ionic-strength effects of protein stability: acid denaturation of Sperm Whale Myoglobin. J. Mol. Biol. 1994;237:602–612. doi: 10.1006/jmbi.1994.1258. [DOI] [PubMed] [Google Scholar]
- 45.Franklin J.C., Cafiso D.S. Internal electrostatic potentials in bilayers: measuring and controlling dipole potentials in lipid vesicles. Biophys. J. 1993;65:289–299. doi: 10.1016/S0006-3495(93)81051-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hirono S., Liu Q., Moriguchi I. High correlation between hydrophobic free energy and molecular surface area characterized by electrostatic potential. Chem. Pharm. Bull. (Tokyo) 1991;39:3106–3109. [Google Scholar]
- 47.Rashin A.A., Iofin M., Honig B. Internal cavities and buried waters in globular proteins. Biochemistry. 1991;25:3619–3625. doi: 10.1021/bi00360a021. [DOI] [PubMed] [Google Scholar]
- 48.Welinder K.G., Bjornholm B., Dunford H.B. Functions of electrostatic potentials and conserved distal and proximal His- Asp H-bonding networks in haem peroxidases. Biochem. Soc. Trans. 1995;23:257–262. doi: 10.1042/bst0230257. [DOI] [PubMed] [Google Scholar]
- 49.Craescu C.T., Schaeffer C., Mispelter J., Garin J., Rosa J. High-resolution NMR studies of histidine-substituted and histidine-perturbed hemoglobin variants. Histidine assignments, electrostatic interactions at the protein surface, and implications for hemoglobin S polymerization. J. Biol. Chem. 1986;261:7894–7901. [PubMed] [Google Scholar]
- 50.Cocco M.J., Kao Y.H., Phillips A.T., Lecomte J.T. Structural comparison of apomyoglobin and metaquomyoglobin: pH titration of histidines by NMR spectroscopy. Biochemistry. 1992;31:6481–6491. doi: 10.1021/bi00143a018. [DOI] [PubMed] [Google Scholar]
- 51.Nadolny C., Kempf I., Zundel G. Specific interactions of the allosteric effector 2,3-bisphosphoglycerate with human hemoglobin—a difference FTIR study. Biol. Chem. Hoppe Seyler. 1993;374:403–407. doi: 10.1515/bchm3.1993.374.1-6.403. [DOI] [PubMed] [Google Scholar]
- 52.Honore B., Brodersen R. Ionization of Tyrosine residues in human serum albumin and in its complexes with bilirubin and laurate. Int. J. Pept. Protein Res. 1992;39:24–28. doi: 10.1111/j.1399-3011.1992.tb01551.x. [DOI] [PubMed] [Google Scholar]
- 53.Sham Y.Y., Chu Z.T., Warshel A. Consistent calculations of pKa of ionizable residues in proteins—semimicroscopic and microscopic approaches. J. Phys. Chem. B. 1997;101:4458–4472. [Google Scholar]
- 54.Davoodi J., Wakarchuk W.W., Campbell R.L., Carey P.R., Surewicz W.K. Abnormally high pKa of an active-site glutamic acid residue in Bacillus circulans xylanase. The role of electrostatic interactions. Eur. J. Biochem. 1995;323:839–843. [PubMed] [Google Scholar]
- 55.Wade R.C., Goodford P.J. The role of hydrogen bonds in drug binding. Prog. Clin. Biol. Res. 1989;289:433–444. [PubMed] [Google Scholar]
- 56.Zhang T., Koshland D.E., Jr Computational method for relative binding energies of enzyme–substrate complexes. Protein Sci. 1997;5:348–356. doi: 10.1002/pro.5560050219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Akke M., Forsen S. Protein stability and electrostatic interactions between solvent exposed charged side chains. Proteins. 1990;8:23–29. doi: 10.1002/prot.340080106. [DOI] [PubMed] [Google Scholar]
- 58.Swartz P.D., Beck B.W., Ichiye T. Structural origin of redox potentials in Fe-S proteins—electrostatic potentials of crystal structures. Biophys. J. 1996;71:2958–2969. doi: 10.1016/S0006-3495(96)79533-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lokhart D.J., Kim P.S. Internal Stark effect measurements of the electric field at the amino terminus of a α-helix. Science. 1992;257:947–951. doi: 10.1126/science.1502559. [DOI] [PubMed] [Google Scholar]
- 60.Friedrich K., Wooley P. Electrostatic potential of macromolecules measured by pKa shift of a fluorophore. I. The 3′ terminus of 16 S RNA. Eur. J. Biochem. 1988;173:227–231. doi: 10.1111/j.1432-1033.1988.tb13988.x. [DOI] [PubMed] [Google Scholar]
- 61.Makhatadze G.I., Loladze V.V., Ermolenko D.N., Chen XF., Thomas S.T. Contribution of surface salt bridges to protein stability: guidelines for protein engineering. J. Mol. Biol. 2003;327:1135–1148. doi: 10.1016/s0022-2836(03)00233-x. [DOI] [PubMed] [Google Scholar]
- 62.Makhatadze G.I., Loladze V.V., Gribenko A.V., Lopez M.M. Mechanism of thermostabilization in a designed cold shock protein with optimized surface electrostatic interactions. J. Mol. Biol. 2004;336:929–942. doi: 10.1016/j.jmb.2003.12.058. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


