Abstract
PAST is a new web service providing fast structural queries of the Protein Data Bank. The search engine is based on an adaptation of the generalized suffix tree and relies on a translation- and rotation-invariant representation of the protein backbone. The search procedure is completely independent of the amino acid sequence of the polypeptide chains. The web service works best with, but is not necessarily limited to, shorter fragments such as functional motifs—a task that most other tools do not perform well. Usual query times are in the order of seconds, allowing a truly interactive use. Unlike most established tools, PAST does not prefilter the dataset or exclude parts of the search space based on statistical reasoning. The server is freely available at http://past.in.tum.de/.
INTRODUCTION
As the Protein Data Bank (PDB) (1) today (January 2006) holds >30 000 structures and continues to grow by >100 structures per week, fast and effective methods for identifying similarities between proteins are needed. While proven and efficient methods based on the comparison of the (amino acid) sequence exist, similar tools for the better preserved structural similarities have thus far been lacking. Computationally, searching in large geometrical databases is a difficult problem. With respect to this problem, established approaches ‘fail’ in the following ways:
They try to sidestep the challenge of many geometrical comparisons by prefiltering the candidate set according to various heuristics, e.g. according to amino acid sequence similarity (which can miss matches because of worse sequence conservation versus conservation of structure), or they work only with a small subset of all PDB entries.
Most existing methods are based on exhaustive search and pairwise comparison, which leads, as a consequence to query times scaling at least linearly with the numbers of structures to be searched. Owing to the nearly exponential growth in number of structures in the PDB, this must be considered unsatisfactory.
Existing tools are too slow for interactive exploration of the currently existing protein data. A typical search for a protein substructure of interest can take minutes to hours or even days.
We found these shortcomings in the following tools: DALI (1), CE (3), PRIDE (4), VAST (5), SPASM (6), SSM (7), TOPS (8) and YAKUSA (9), among others. We will show that it is possible to perform true interactive searches for identical or similar backbone conformations of protein (sub)structures. Our approach, PAST (Polypeptide Angle Suffix Tree), first described in Ref. (10), uses a linear representation of the protein backbone conformation which is stored in a suffix tree. Unlike most other search methods, PAST does not have to use a filtered subset of ‘structural representatives’ but includes all polypeptide structures contained in the PDB.
METHODS
Protein structure representation
To describe the 3D conformation of a protein we use the sequence of α, the dihedral torsion angle around the virtual bonds between two consecutive Cα atoms defined by the four Cα atoms i − 1, i, i + 1 and i + 2. These torsion angles have the advantage of being invariant to translation and rotation of the protein structure in the actual coordinate system.
The angles are encoded into an alphabet (represented by the characters with ASCII codes from 1 to 36) by discretizing in intervals of size 360°/36 = 10°. This transforms the information of the 3D backbone conformation from all protein structures contained in the PDB into sequences of ‘structural texts’ which are then stored in the suffix tree data structure. An alternative encoding using the sequence of the backbone dihedral torsion angles φ and ψ will be added in the near future. (It is already available for the local installation version.)
Construction of PAST
For the initial construction of the indexing data structure, an extension of Ukkonen's algorithm (11) to generalized suffix trees (12) is used. For PDB files containing polypeptide structures, all chains, models and alternative location indicators are handled as separate entries and included in PAST. As of January 2006, a total of 32 800 files describing polypeptide structures gives 165 000 separate entries in PAST. The computation of the generalized suffix tree, given the α angle sequences of all polypeptide entries of the PDB, takes ∼2–3 min on a standard PC (1 GHz). The size of the suffix tree is <2 GB; thus it can be held in main memory, making all calculations extremely fast.
Exact and approximate matching
Exact matching is performed by computing the respective dihedral torsion angles of the query structure, encoding the angles into characters analogously to the database preprocessing step and using the resulting text to perform a suffix tree search. However, in most cases one wants to find not only identical matches but also entries similar to the query structure. Approximate matching is performed by including ‘neighboring’ characters (i.e. neighboring torsion angle intervals) in the search procedure. The worst case query time complexity of the exact search method does not depend on the size of the database. It is bounded by a linear function of query sequence length and the number of occurrences (hits). The approximate search is still very fast in practice because the resulting suffix tree is sparse compared to the angle sequence space, even though its worst case time complexity is much worse in theory (exponential in the length of the search pattern). Fora more detailed description of the data structures and algorithms used see Ref. (12).
Post-processing
Post-processing of the results set includes root mean square deviation (r.m.s.d.) calculation of values between Cα atoms of the protein backbone structure from the query and all matching database entries. Despite a quality measure for structural similarity of the hits, the calculated Cα r.m.s.d. value is used as a cutoff for the resulting output.
USAGE
Querying PAST
In order to perform a search for equal or similar protein backbone conformations in the PDB the web interface offers the following options:
PDB id/file: Give the four-letter PDB ID of the query structure (e.g. 1MFS) or upload, a local file containing the query structure coordinates in PDB format.
Model/Chain/Alternate location: Specify the PDB MODEL number, Chain ID and Alternate Location Indicator.
First/Last residue: Give the atom track numbering of the first and last residue of the query segment (e.g. 15/28). Since this search method relies on a continuous linear description of the protein backbone, the specified query segments should have a complete atom track record. If this requirement is not met, the computed torsion angles must be regarded as unreliable. Of course, the same holds true for the sequences of potential target structures. By testing our software we had noticed that not all PDB entries meet this requirement. With the last three entries the search parameters have to be specified:
Angle type: Search based on the virtual bond dihedral torsion angles (α).
Tolerance: Range of neighboring angle intervals regarded as hits (respectively, for every position).
r.m.s.d. cut-off: Displays only matches with a Cα coordinates r.m.s.d. below or equal to the given value (post-processing).
A tolerance of ±0 allows for all positions only the exact matches within the original 10 coding interval and ±k allows additional k intervals to both sides of the original query interval at all positions. The tolerance should be started with small intervals (1–3) and increased successively up to 10–12, until the result set becomes too unspecific. The optimal values for tolerance and r.m.s.d. cut-off depends on the size and structural conservation of the query and of course the degree of structural similarity that an user regards as a ‘true hit’. The r.m.s.d. cut-off can be set to rather high values if no filtering is wanted (e.g. 15 Å).
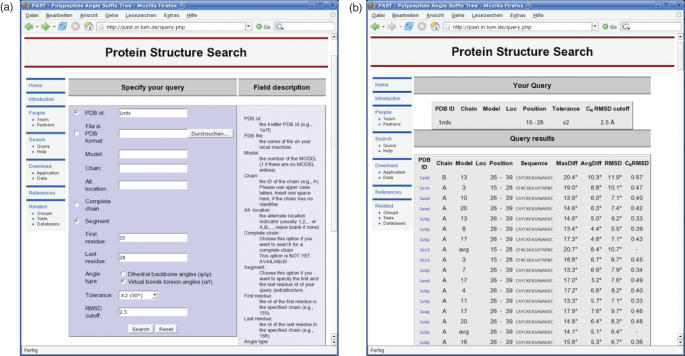
‘Fine tuning’ of searches can easily be performed by iterating from the results table back to the query window by using the ‘Back’ button of the browser, successive modification of the search parameters and re-submitting the new query. Owing to space constraints, a short tutorial that shows the typical use of this web server is provided in the online Supplementary Data. A screenshot of the query interface is shown in Figure 1a.
Figure 1.
Screenshots of PAST running the example query.
Interpreting the results
After submission of a query the results table should be loaded automatically within a short span of time. A screenshot of the results table for the test example is shown in Figure 1b. The results page is structured as follows: on the top of the table the search parameters are given followed by an (unsorted) list of all (sub)structures that match the respective query. For each fragment, the PDB ID, Model, Chain and Alternate Location Indicator (Loc) together with the Position (first and last residues) and the respective amino acid sequence is given. The PDB IDs are linked to the respective PDB entry files. The last column of the table shows the search specific calculation results. The degree values are PAST specific owing to the internal torsion angle representation of the structures.
MaxDiff shows the maximum occurring angle difference between the query and the match.
AvgDiff shows the average angle difference between the query and the match.
r.m.s.d. gives the squared r.m.s.d. values of the angle differences between the query and the match.
Cα r.m.s.d. gives the squared r.m.s.d. values of Cα atom coordinates of the query and matching polypeptide structure in Å.
The Cα r.m.s.d. offers a algorithm/approach independent similarity measure for the found matches. Note that the Cα r.m.s.d. is calculated including all Cα atoms of the query and target segment.
Example query
The following example query uses the CCHC zinc finger motif of the PDB entry 1MFS (residues 15–28). Searching with the α torsion angles by using a tolerance of ±0 coding intervals (exact matching) and a Cα r.m.s.d. cut-off at 2.5 Å only the respective PDB entry itself is found. Raising the tolerance to ±2 leads to 308 matching structures belonging to 15 different PDB entries (data not shown here in detail). All models and calculated averages of a PDB entry are shown as separate matches. With an quick inspection of the results that are using the provided amino acid sequence all entries can easily be identified as members of the respective SCOP (Retrovirus zinc finger-like domain) and PROSITE (PS50158) families. Without Cα r.m.s.d. filtering, the first ‘false positive’ hit (1CJG, MaxDiff 28.8, Cα r.m.s.d. 3.08 Å) occurs using a tolerance of ±3 intervals, which means a total search range of seven coding intervals (i.e. 70 allowed for each position).
CONCLUSION
Our method of discretizing the backbone torsion angles and putting the respective character encoding into a generalized suffix tree has proven to be a very fast solution for answering queries about local structural similarities to the PDB. As the web server implementation of PAST performs an ungapped structural alignment using an overall maximum deviation cut-off, its strength lies in the identification of short polypeptide fragments of local similarity rather than comparingfull protein chains. Queries of ∼10–50 residues gave good results during the testing phase. Most established tools for structural comparison (exept SPASM) either perform badly on such short query structures (SSM, CE, VAST) or even do not even accept them like DALI.
Compared with SPASM, PAST shows comparable or better results, while beeing much faster. More detailed results are provided in the Supplementary Data and are also given in Ref. (13).
Implemented in the web service PAST, our method, improves on the shortcomings mentioned in the Introduction in the following ways:
Searches are performed including all polypeptide structures contained in the PDB.
It is based on the data structure of a suffix tree and hence shares an interesting property with it: the search time does not depend on the size of the database, but on the length of the query structure and the number of matches.
On our web server the usual search time is in the order of seconds, enabling true interactive working by repeated searches. At the same time, the quality of the results is at least comparable to established structural search tools.
Hence, we consider PAST to be a valuable tool for the fast detection of short consecutive protein backbone structures usually found in motifs and domains.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
Acknowledgments
We appreciate the support by Prof. E. W. Mayr. We al thank Moritz Maaß for valuable suggestions concerning the suffix tree implementation and analysis. Special thanks go the Stella Clarke for proof reading our concepts. Further we want to acknowledge the support by Jürgen Paal from ALTANA Pharma. As well as the improved search functions by Uwe Römers and Anselm Kusser. We al thank the anonymous reviewers for helpful considerations. Funding to pay the Open Access publication charges for this article was provided by xxxxx.
Conflict of interest statement. None declared.
REFERENCES
- 1.Berman H.M., Westbrook J.D., Feng Z., Gilliland G.L., Bhat T.N., Weissig H., Shindyalov I.N., Bourne P.E. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Holm L., Sander C. Searching protein structure databases has come of age. Proteins. 1994;19:165–173. doi: 10.1002/prot.340190302. [DOI] [PubMed] [Google Scholar]
- 3.Shindyalov I.N., Bourne P.E. Protein structure alignment by incremental combinatorial extension (CE) of the optimal path. Protein Eng. 1998;11:739–747. doi: 10.1093/protein/11.9.739. [DOI] [PubMed] [Google Scholar]
- 4.Gáspári Z., Vlahovicek K., Pongor S. Efficient recognition of folds in protein 3D structures by the improved PRIDE algorithm. Bioinformatics. 2005;21:3322–3323. doi: 10.1093/bioinformatics/bti513. [DOI] [PubMed] [Google Scholar]
- 5.Gibrat J.-F., Madej T., Bryant S.H. Surprising similarities in structure comparison. Curr. Opin. Struct. Biol. 1996;6:377–385. doi: 10.1016/s0959-440x(96)80058-3. [DOI] [PubMed] [Google Scholar]
- 6.Kleywegt G.J. Recognition of spatial motifs in protein structures. J. Mol. Biol. 1999;285:1887–1897. doi: 10.1006/jmbi.1998.2393. [DOI] [PubMed] [Google Scholar]
- 7.Krissinel E., Henrick K. Secondary-structure matching (SSM), a new tool for fast protein structure alignment in three dimensions. Acta Crystallogr. 2004;D60:2256–2268. doi: 10.1107/S0907444904026460. [DOI] [PubMed] [Google Scholar]
- 8.Gilbert D.R., Westhead D.R., Nagano N., Thornton J.M. Motif-based searching in TOPS protein topology databases. Bioinformatics. 1999;15:317–326. doi: 10.1093/bioinformatics/15.4.317. [DOI] [PubMed] [Google Scholar]
- 9.Carpentier M., Brouillet S., Pothier J. YAKUSA: a fast structural database scanning method. Proteins. 2005;61:137–151. doi: 10.1002/prot.20517. [DOI] [PubMed] [Google Scholar]
- 10.Buchner A., Täubig H. A fast method for motif detection and searching in a protein structure database. 2003. Technical Report TUM-I0314, Inst. f. Informatik, TU München.
- 11.Ukkonen E. On-line construction of suffix trees. Algorithmica. 1995;14:249–260. [Google Scholar]
- 12.Bieganski P., Riedl J., Carlis J.V., Retzel E.F. Generalized suffix trees for biological sequence data: applications and implementation. Proceedings of the 27th Annual Hawaii International Conference on System Sciences (HICSS'94), Vol. V (Biotechnology Computing); IEEE; 1994. pp. 35–44. [Google Scholar]
- 13.Täubig H., Buchner A., Griebsch J. A method for fast approximate searching of polypeptide structures in the PDB. Proceedings of the German Conference on Bioinformatics (GCB'04), Vol. P-53 of Lecture Notes in Informatics; Köllen Verlag; 2004. pp. 65–74. [Google Scholar]