Abstract
DNA vaccines have slowly emerged as keystones in preventive immunology due to their versatility in inducing both cell-mediated as well as humoral immune responses. The design of an efficient DNA vaccine, involves choice of a suitable expression vector, ensuring optimal expression by codon optimization, engineering CpG motifs for enhancing immune responses and providing additional sequence signals for efficient translation. DyNAVacS is a web-based tool created for rapid and easy design of DNA vaccines. It follows a step-wise design flow, which guides the user through the various sequential steps in the design of the vaccine. Further, it allows restriction enzyme mapping, design of primers spanning user specified sequences and provides information regarding the vectors currently used for generation of DNA vaccines. The web version uses Apache HTTP server. The interface was written in HTML and utilizes the Common Gateway Interface scripts written in PERL for functionality. DyNAVacS is an integrated tool consisting of user-friendly programs, which require minimal information from the user. The software is available free of cost, as a web based application at URL: http://miracle.igib.res.in/dynavac/.
BACKGROUND
DNA Vaccines have been one of the latest developments in vaccine technology. DNA vaccines are essentially plasmids capable of expressing an antigenic peptide in the host. DNA vaccines have become an attractive alternative to conventional methods due to the fact that it can elicit sustained cell mediated as well as humoral immune responses, which is very much important in combating pathogenic organisms, especially intracellular pathogens.
Recently several techniques like optimizing codons (1), engineering CpG motifs (2,3), introducing promoter sequences (4–6) and co-injection with plasmids expressing immunostimulatory molecules have been tried successfully to enhance the immunogenicity of DNA vaccines. These sequential and inter-dependent changes to be performed to create optimal vaccines make vaccine design a largely unsystematic and tedious process.
Here we report DyNAVacS, an integrative bioinformatics tool for optimized DNA vaccine design. The software is designed to be modular and assists the user through a systematic and stepwise process of vaccine design thus making it user-friendly. This software can also be used for optimizing codons for heterologous expression of genes in bacteria, yeast and plants, mapping restriction enzyme sites, primer design and design of genes for gene therapy, thus providing a complete repertoire of tools for cloning and expression of proteins.
MATERIALS AND METHODS
Codon usage tables
The codon usage tables were derived from the Kazusa Codon Usage Database (http://www.kazusa.or.jp/codon/), which provides codons and the frequencies in their respective genomes.
Identification of highly expressed genes
Genome-wide expression profiling data was used for determining the set of highly expressed genes in various genomes. Gene expression data from high density oligonucleotide array based experiments for plants, yeast and mouse were derived from repositories for microarray data (7,8) or previous reports (9), Control data were retrieved from Affymetrix datasets submitted from different laboratories in public repositories like TAIR and yMGV. The fluorescence intensity was linearized by log transformation and Z-values were calculated to normalise the data relative to the mean and in terms of standard deviation (10).
Z-values were calculated using the formula
Average of Z-values for each gene were calculated and genes with consistently high Z-scores over at least four independent experiments were designated highly expressed genes. The sequences were then retrieved from GenBank and codon usage tables were derived using codon count (http://www.kazusa.or.jp/codon/) program. In the case of mouse, the coding sequences of genes whose promoters have been used for high expression were used for generating the codon usage table.
Vectors
Data regarding vectors that had been previously used for transgenic expression were collected from literature.
WEB APPLICATION
DyNAVacS utilizes Apache as web server and CGI (Common Gateway Interface) scripts written in PERL is used to input and output data from and to an HTML interface. The tool is freely available at the URL http://miracle.igib.res.in/dynavac/. The flowchart at the right end of the homepage is also a clickable link to directly proceed to the CpG optimization and other standalone tools. The result tracker allows retrieval of results of previous projects by submitting the project ID.
Codon optimization
DyNAVacS computes the optimal codon for each amino acid encoded by the stretch of DNA and replaces the codons with the most optimal codon according to the codon usage tables derived from the Kazusa Codon Usage Database (http://kazusa.or.jp/codon). Codon usage tables for mouse, Arabidopsis and yeast specially prepared from highly expressed genes can also be chosen. The codon usage tables for most commonly used expression systems including marine organisms can be selected.
Engineering CpG motifs
Two alternative methods are provided for optimization of CpG content in the sequence. The first method optimizes the codons for CG dinucleotides, provided the usage frequency is not <80% of the maximum, thus providing a fair balance between CG dinucleotide frequency and codon optimization.
The second method inserts the commonly used consensus motif XCGY (where X is any base but C, and Y is any base but G) in the sequence.
DyNAVacS also interactively submits the sequences to the CpGplot server for comparison. A tool for mapping experimentally proven stimulating and suppressive CpG-motifs (11–13) has been incorporated. Di-nucleotide counts for input and output sequences can also be obtained.
Restriction enzyme site mapping
DyNAVacS can identify and map restriction sites corresponding to 7504 restriction Enzymes.
Primer selection
A simple primer design tool is provided for quick PCR applications wherein the melting temperature of the primer is calculated using the formula:
where length is the length of the primer, [Na+] is the salt concentration and %GC is the percentage of GC-content.
The Primer3 algorithm has been incorporated for more advanced primer design applications (14).
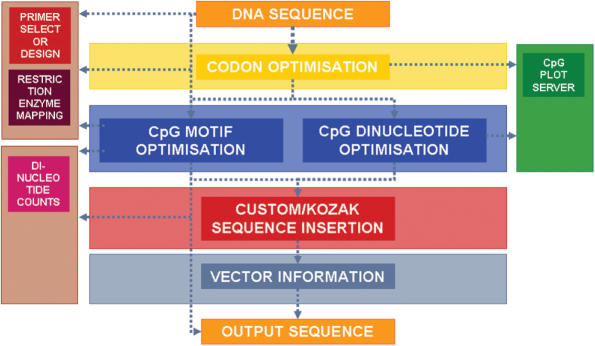
An overview of the organization and functionality of the modules is depicted in Figure 1.
Figure 1.
Flowchart depicting the organization and functionality of different modules in DyNAVacS.
RESULTS AND DISCUSSION
DyNAVacS is designed to be modular, with individual modules integrated using CGI scripts coded in PERL. A brief description of each module is summarized in Table 1.
Table 1.
Summary of modules incorporated in DyNAVacS
| Modules available | Functionality of the tool |
|---|---|
| Codon optimization | The software optimizes the input sequence by recognizing the optimal codon corresponding to each amino acid from codon usage table. A list of codon usage tables based on Kazusa Codon Usage Database and highly expressed genes is provided to users to select the host organism |
| CpG optimization | Engineering CpG motifs in the optimized sequence. This is done by two methods: by incorporating general motif of CpG motifs XCGY and by striking a balance between G and C content and frequency of codon usage. User specifies the start and the end positions of the CpG motif |
| Restriction enzyme site mapping | The software can map sites corresponding 7504 type II restriction enzymes |
| Kozak sequence insertion | Insertion of Kozak sequences chosen by user in the CpG optimized sequence |
| Custom sequence insertion | This option gives the user freedom of inserting any sequence into CpG optimized sequence at the position specified by user. |
| Information on vectors | The user can obtain appropriate information on vectors from the table provided containing properties of vectors like vector name, promoter, enhancer, terminator, selective markers, etc. |
| Primer select/design | This module enables the user to select primers from the sequences at any intermediate stage. Tm and 3′ inter-primer complementarity can also be obtained. The Primer3 algorithm can be used for more extensive applications |
The interface of DyNAVacS is user-friendly and comes with a host of logical options at each step and the user can have the flexibility to go back and redo any step at any point (Figure 2). The user can also have the advantage of comparing his outputs interactively, e.g. the CpG optimizations on the CpG plot server at EBI. However, the process of codon/CpG optimization may inadvertently give rise to novel structural determinants splice sites etc, which are not addressed in the current version.
Figure 2.
Input page of DyNAVacS.
The primer select tool is meant for selecting potentially useful regions for primer design and checking physical parameters like melting temperature to aid in choice of primers.
To the best of our knowledge, there are no other servers which integrate the various steps involved in DNA vaccine design. A standalone tool Upgene (15) carries out codon optimization through a randomization-based codon replacement approach. The tool however has not been applied extensively by experimentalists.
CONCLUSIONS
DyNAVacS integrates a host of experimentally validated tools in a systematic and stepwise manner to design DNA vaccines optimized for antigenicity as well as heterologous expression. In addition to vaccine design, the modular design of DyNAVacS enables it to be useful in areas like gene therapy and design of synthetic genes for heterologous expression.
AUTHORS' CONTRIBUTIONS
N.H. and R.G. coded the basic modules of the software. P.A. and B.P. compiled the data on highly expressed genes. V.S. coded the web interface and integrated the software. N.H., R.G., V.S. and B.P. wrote the manuscript.
Acknowledgments
The authors acknowledge S. K. Brahmachari and P. Burma for discussions, JNCASR for fellowships to N.H. through the SRFP-2005 and CSIR India for funding. The authors would also like to thank Manoj Hariharan for invaluable inputs and suggestions. The Open Access publication charges for this article were waived by Oxford University Press.
Conflict of interest statement. None declared.
REFERENCES
- 1.Uchijima M., Yoshida A., Nagata T., Koide Y. Optimization of codon usage of plasmid DNA vaccine is required for the effective MHC class I-restricted T cell responses against an intracellular bacterium. J. Immunol. 1998;161:5594–5599. [PubMed] [Google Scholar]
- 2.Klinman D.M., Yamshchikov G., Ishigatsubo Y. Contribution of CpG motifs to the immunogenicity of DNA vaccines. J. Immunol. 1997;158:3635–3639. [PubMed] [Google Scholar]
- 3.Krieg A.M., Yi A.K., Schorr J., Davis H.L. The role of CpG dinucleotides in DNA vaccines. Trends Microbiol. 1998;6:23–27. doi: 10.1016/S0966-842X(97)01145-1. [DOI] [PubMed] [Google Scholar]
- 4.Mukhtar M., Duan L., Bagasra O., Pomerantz R.J. Evaluation of relative promoter strengths of the HIV-1-LTR and a chimeric RSV-LTR in T lymphocytic cells and peripheral blood mononuclear cells: promoters for anti-HIV-1 gene therapies. Gene Ther. 1996;3:725–730. [PubMed] [Google Scholar]
- 5.Lee A.H., Suh Y.S., Sung J.H., Yang S.H., Sung Y.C. Comparison of various expression plasmids for the induction of immune response by DNA immunization. Mol. Cells. 1997;7:495–501. [PubMed] [Google Scholar]
- 6.Xu Z.L., Mizuguchi H., Ishii-Watabe A., Uchida E., Mayumi T., Hayakawa T. Optimization of transcriptional regulatory elements for constructing plasmid vectors. Gene. 2001;272:149–156. doi: 10.1016/s0378-1119(01)00550-9. [DOI] [PubMed] [Google Scholar]
- 7.Barrett T., Suzek T.O., Troup D.B., Wilhite S.E., Ngau W.C., Ledoux P., Rudnev D., Lash A.E., Fujibuchi W., Edgar R. NCBI GEO: mining millions of expression profiles—database and tools. Nucleic Acids Res. 2005;33:D562–D566. doi: 10.1093/nar/gki022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Le Crom S., Devaux F., Jacq C., Marc P. yMGV: helping biologists with yeast microarray data mining. Nucleic Acids Res. 2002;30:76–79. doi: 10.1093/nar/30.1.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sharp P.M., Li W.H. Codon usage in regulatory genes in Escherichia coli does not reflect selection for ‘rare’ codons. Nucleic Acids Res. 1986;14:7737–7749. doi: 10.1093/nar/14.19.7737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cheadle C., Vawter M.P., Freed W.J., Becker K.G. Analysis of microarray data using Z-score transformation. J. Mol. Diagn. 2003;5:73–81. doi: 10.1016/S1525-1578(10)60455-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bauer M., Redecke V., Ellwart J.W., Scherer B., Kremer J.P., Wagner H., Lipford G.B. Bacterial CpG-DNA triggers activation and maturation of human CD11c-, CD123+ dendritic cells. J. Immunol. 2001;166:5000–5007. doi: 10.4049/jimmunol.166.8.5000. [DOI] [PubMed] [Google Scholar]
- 12.Krieg A.M., Yi A.K., Matson S., Waldschmidt T.J., Bishop G.A., Teasdale R., Koretzky G.A., Klinman D.M. CpG motifs in bacterial DNA trigger direct B-cell activation. Nature. 1995;374:546–549. doi: 10.1038/374546a0. [DOI] [PubMed] [Google Scholar]
- 13.Van Uden J., Raz E. Introduction to immunostimulatory DNA sequences. Springer Semin. Immunopathol. 2000;22:1–9. doi: 10.1007/s002810050010. [DOI] [PubMed] [Google Scholar]
- 14.Rozen S., Skaletsky H. Primer3 on the WWW for general users and for biologist programmers. Methods Mol. Biol. 2000;132:365–86. doi: 10.1385/1-59259-192-2:365. [DOI] [PubMed] [Google Scholar]
- 15.Gao W., Rzewski A., Sun H., Robbins P.D., Gambotto A. UpGene: Application of a web-based DNA codon optimization algorithm. Biotechnol. Prog. 2004;20:443–448. doi: 10.1021/bp0300467. [DOI] [PubMed] [Google Scholar]