Abstract
A variety of methods is used for a molecular typing of Enterococcus spp. and related gram-positive bacteria including macrorestriction analysis using pulsed-field gel electrophoresis (PFGE), ribotyping, rapid amplification of polymorphic DNA (RAPD), and amplified fragment length polymorphism (AFLP). To test the influence of transferable determinants on the outcome of different typing methods commonly used for enterococci, we established a homogenous strain collection of 24 transconjugants resulting from filter matings with antibiotic-resistant Enterococcus faecium. As expected, AFLP, RAPD, and PFGE all identified our model bacteria as strongly related. However, distinct differences in the resolving and discriminatory power of the tested methods could be clearly addressed. In PFGE, 22 of 24 transconjugants possessed less than a three-band difference to the recipient pattern and would be regarded as strongly related. Three different RAPD PCRs were tested; in two reactions, identical patterns for all transconjugants and the recipient were produced. One RAPD PCR produced an identical pattern for 18 transconjugants and the recipient and a clearly different pattern for the remaining 6 transconjugants due to a newly appearing fragment resulting from acquisition of the tetL gene. AFLP clusters all transconjugants into a group of major relatedness. Percent similarities were highly dependent on the method used for calculating the similarity coefficient (curve-based versus band-based similarity coefficient). Fragment patterns of digested plasmids showed the possession of nonidentical plasmids in most transconjugants. PFGE still could be recommended as the method of choice. Nevertheless, the more-modern AFLP approach produces patterns of comparable discriminatory power while possessing some advantages over PFGE (less-time-consuming internal standards). Plasmid fingerprints can be included to subdifferentiate enterococcal isolates possessing identical macrorestriction and PCR typing patterns.
The “gold standard” for molecular typing of enterococci and related gram-positive bacteria, such as staphylococci and lactococci is still macrorestriction analysis via pulsed-field gel electrophoresis (PFGE) (13, 14). In recent years, some alternative techniques have been successfully applied to the typing of enterococci below the species level. These include amplification-based methods, such as rapid amplification of polymorphic DNA (RAPD) (16, 26) and amplified fragment length polymorphisms (AFLP) (2, 19). These techniques are now applied more and more because they involve less time, comparably low costs, and only standard equipment. Fragments resulting from RAPD PCR typing are randomly amplified and resolved in common agarose gels. AFLP is used for a wide range of organisms with different applications and has been successfully applied to the study of the genetic relatedness of epidemiological unrelated strains of enterococci (33). AFLP-resulting fragments are determined in a capillary or gel sequencer allowing detection of single base pair differences in the corresponding fragment patterns. In addition to AFLP, multilocus sequence typing is a relatively new technique which has been applied to the study of the global epidemiology of different bacterial pathogens (9), including the recent description of a scheme for Enterococcus faecium (11).
Amplification-based methods such as RAPD and AFLP are often used empirically. Without knowledge of the complete genome sequence (which was and still is the case for most applications), it is not possible to predict what bands should appear in fragment patterns and what fragments are nonspecific. In general, these methods work quite well when compared to already established methods, such as PFGE or ribotyping. However, it is not known to what extent major and minor molecular events in a cell, such as rearrangements of chromosomal fragments, point mutations, and the presence of plasmids, influence the outcome of amplification-based typing schemes. To evaluate the influence of transferable markers (e.g., plasmids) on the outcome of genotyping and to test the discriminatory power of the most commonly used methods for typing of enterococci, we chose a quite different approach. A homogenous strain collection was generated by filter-mating transfer of genetic determinants from 24 multiple-antibiotic-resistant E. faecium isolates into a recipient isolate while selecting for transfer of antibiotic resistance determinants. The influence of transferred determinants on the outcome of PFGE, RAPD, and AFLP typing was investigated and compared with restriction patterns of digested plasmids.
MATERIALS AND METHODS
Strains, media, antibiotic susceptibilities.
Antibiotic-resistant strains from our strain collection originated from already published studies (28, 29, 30, 31). They were known to transfer resistance determinants by filter mating (28, 29). Strain no. 64 isolated from a stool sample of a hospital patient was susceptible to all antibiotics and free of plasmids. A rifampin- and fusidic acid-resistant derivative, 64/3, was selected by spontaneous mutation and was used as the recipient, R, in filter matings (12, 28). Isolates were grown on brain heart infusion (BHI) agar or in BHI broth (Difco Labs., Detroit, Mich.).
Antibiotic susceptibilities and filter mating.
All MIC tests were done by microbroth dilution in Isosensitest broth as described elsewhere (29), following the instructions of the DIN 58940 (German Institute for Standards). Reference ranges for susceptibility (s) and resistance (r) are as follows (in micrograms per milliliter): glycopeptides, s ≤ 4, r ≥ 16; penicillin, s ≤ 8, r ≥ 16; ampicillin, s ≤ 2, r ≥ 16; streptomycin, s ≤ 8, r ≥ 32; erythromycin and tetracycline, s ≤ 1, r ≥ 8; chloramphenicol, s ≤ 8, r ≥ 16; ciprofloxacin, s ≤ 0.25, r ≥ 2; trimethoprim-sulfamerazine, s ≤ 4, r ≥ 32; rifampin, s ≤ 0.5, r ≥ 1; fusidic acid, s ≤ 2, r ≥ 4; mupirocin, s ≤ 2, r ≥ 32; and quinupristin-dalfopristin (Synercid), s ≤ 2, r ≥ 4 (29). Filter mating was done on nitrocellulose filters (0.45-μm pore size; Sartorius, Goettingen, Germany) placed on BHI agar. Exponentially growing donor and recipient cells (1 ml of each) were placed together on a filter and incubated for 4 h or overnight at 37°C. Transconjugants were selected on BHI agar supplemented with rifampin (30 μg/ml) and the second selectable marker. Transconjugants were separated on agar containing the selective marker for the donor and the nonselective marker from the recipient (fusidic acid, 20 μg/ml).
Molecular methods.
Preparation of samples and subsequent macrorestriction analysis was done in a CHEF III apparatus (BIO-RAD, Munich, Germany) as already described (29) with the following modifications: agarose gel concentration was 1% and ramped pulse times were 1 to 11 s for 13 h and 11 to 30 s for 13 h. Bacterial DNA fingerprints were analyzed by molecular mass determination of all DNA fragments with Staphylococcus aureus strain NCTC 8325 as a reference. Similarities among fingerprinting patterns were estimated by eye and by a band-based similarity coefficient (Dice) according to the method of Claus et al. (8).
DNA for amplification-based techniques was isolated by using a DNeasy tissue kit from Qiagen (Hilden, Germany). The DNA was stained with an intercalating fluorescent dye from the Pico Green kit (Molecular Probes Europe, Leiden, The Netherlands) and concentration was measured in a fluorimager FLA-2000 (Raytest Germany, Straubenhardt, Germany). The appropriate concentration was estimated for RAPD and AFLP typing. The protocol for AFLP was followed exactly as published recently in detail (33). The percent similarity of AFLP-generated patterns was calculated with both a curve-based similarity coefficient (Pearson product moment correlation) and a band-based binary similarity coefficient (Dice, BioNumerics software, version 2.5; Applied Maths, Sint-Martens-Latem, Belgium). The unweighted pair group method with arithmetic averages (UPGMA) was used to cluster the matrix of similarity coefficients (Applied Maths).
PCR was performed by using RAPD analysis beads from Amersham Biosciences Europe GmbH (Freiburg, Germany). The DNA concentration was 50 ng per PCR, and primers were diluted to a final concentration of 1 μM. Three primers were tested: 5′-TCCCGCGCA (16) (RAPD3), 5′-TGCTCTGCCC (RAPD1), and 5′-GTAGACCCGT (26) (RAPD2). The PCR conditions for primer RAPD3 were as described previously (16). The PCR conditions for primers RAPD1 and RAPD2 were as follows. After a denaturing step, 35 cycles were run for 30 s at 94°C, 30 s at 34°C, and 60 s at 72°C followed by a final step of 5 min at 72°C. Ten-microliter samples were resolved through a 2% agarose gel, and the runtime was 2 h at 100 V. A PCR for a specific fragment of tetM was done as follows. The primers were tetM1, 5′-GGTGAACATCATAGACACGC, and tetM2, 5′-CTTGTTCGAGTTCCAATGC, the annealing temperature was 55°C, and the fragment size was 401 bp. Amplification of a fragment of tetL was performed with primers tetL1, 5′-TGGTCCTATCTTCTACTCATTC, and tetL2, 5′-TTCCGATTTCGGCAGTAC, the annealing temperature was 53°C, and the size of the product was 385 bp.
DNA from agarose gels was purified by using a commercial kit QIAEX II from Qiagen. Isolated fragments were cloned into pUC18 by using a Sure Clone Ligations kit for cloning PCR fragments with 3′ A overhangs (Amersham Biosciences). DNA was introduced by standard transformation techniques into recipient Escherichiacoli DH5alpha (18). Transformants appeared as white colonies on agar possessing ampicillin (50 μg/ml), isopropyl-β-d-thiogalactopyranoside (0.1 mM), and 5-bromo-4-chloro-3-indolyl-β-d-thiogalactopyranoside (X-Gal, 40 μg/ml).
Plasmid DNA was isolated according to Woodford et al. (34) with modifications described recently (G. Werner, I. Klare, and W. Witte, Letter, Antimicrob. Agents Chemother. 37:2383-2384, 1999). Ten microliters of these preparations was digested overnight with 20 U of endonuclease HindIII. Samples were electrophoresed through 1% agarose gels. Similarities among plasmid fingerprint patterns were estimated by eye and by a band-based similarity coefficient (Dice) according to the method of Claus et al. (8).
RESULTS
Identification of transconjugants.
Transconjugants were generated in 24 filter matings of vancomycin- or streptogramin-resistant E. faecium (donor) isolates and recipient R. The transconjugants were tested separately on agar containing only the antibiotic selecting the selective marker of the donor or the selective (rifampin resistance) and the nonselective marker (fusidic acid resistance) of the recipient. The antibiotic susceptibilities of each transconjugant (one per mating) were determined by microbroth dilution and are given in Table 1. All isolates were resistant to rifampin and fusidic acid. The transconjugants were susceptible to β-lactams and fluoroquinolones. The corresponding determinants are mostly not transferred in matings between E. faecium, indicating the clonal relatedness of the transconjugants with the recipient isolate R.
TABLE 1.
Antibiotic susceptibilities of the 24 transconjugants and recipient Ra
| Strain | MIC (mg/liter) of:
|
PCR result for:
|
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| ERY | CLI | STR | CMP | VAN | TPL | QD | OTE | tetM | tetL | |
| T1 | >8 | >8 | 64 | 16 | ≤1 | ≤1 | >8 | 1 | ND | − |
| T2 | >8 | >8 | 64 | 16 | ≤1 | ≤1 | >8 | 1 | ND | − |
| T3 | >8 | >8 | >64 | ≤4 | >16 | 8 | >8 | 16 | − | + |
| T4 | >8 | >8 | >64 | ≤4 | >16 | 16 | >8 | 16 | − | + |
| T5 | >8 | >8 | 64 | ≤4 | >16 | 4 | 1 | ≤0.5 | ND | − |
| T6 | >8 | >8 | 32 | 8 | >16 | 16 | 1 | ≤0.5 | ND | − |
| T7 | >8 | >8 | >64 | ≤4 | >16 | >16 | >8 | 16 | − | + |
| T8 | >8 | >8 | >64 | ≤4 | >16 | >16 | >8 | 32 | − | + |
| T9 | >8 | >8 | 32 | 8 | >16 | >16 | 1 | ≤0.5 | − | − |
| T10 | 1 | 8 | 16 | ≤4 | >16 | >16 | 1 | ≤0.5 | − | − |
| T11 | >8 | >8 | 16 | ≤4 | >16 | 16 | 2 | ≤0.5 | − | − |
| T12 | >8 | >8 | 32 | ≤4 | >16 | 16 | 2 | ≤0.5 | − | − |
| T13 | >8 | >8 | 16 | 8 | >16 | >16 | 1 | ≤0.5 | − | − |
| T14 | 1 | ≤0.5 | 16 | ≤4 | >16 | 2 | 1 | 1 | − | − |
| T15 | 1 | ≤0.5 | 32 | 16 | >16 | >16 | 1 | 1 | − | − |
| T16 | >8 | >8 | >64 | ≤4 | >16 | >16 | 1 | ≤0.5 | − | − |
| T17 | >8 | >8 | >64 | 8 | ≤1 | ≤1 | >8 | 256 | + | + |
| T18 | >8 | >8 | >64 | 16 | ≤1 | ≤1 | >8 | 256 | + | + |
| T19 | >8 | >8 | >64 | 16 | ≤1 | ≤1 | >8 | 512 | + | + |
| T20 | >8 | >8 | >64 | 8 | ≤1 | ≤1 | >8 | 256 | + | + |
| T21 | >8 | >8 | >64 | 8 | >16 | 4 | >8 | 256 | + | + |
| T22 | >8 | >8 | >64 | 8 | 4 | ≤1 | >8 | 256 | + | + |
| T23 | >8 | >8 | >64 | 8 | 8 | ≤1 | 8 | 8 | − | + |
| T24 | >8 | >8 | >64 | 8 | >16 | 8 | >8 | 512 | + | + |
| R | ≤0.5 | ≤0.5 | 32 | ≤4 | ≤1 | ≤1 | 1 | ≤0.5 | − | − |
All transconjugants were susceptible to penicillin, ampicillin, ciprofloxacin, trimethoprim-sulfamerazine, and mupirocin. All were resistant to fusidic acid and rifampin. ERY, erythromycin CLI, clindamycin; STR, streptomycin; CMP, chloramphenicol; VAN, vancomycin; TPL, teicoplanin; QD, quinupristin-dalfopristin; OTE, Oxytetracycline; ND, not determined; +, present; −, absent.
Macrorestriction analysis by PFGE.
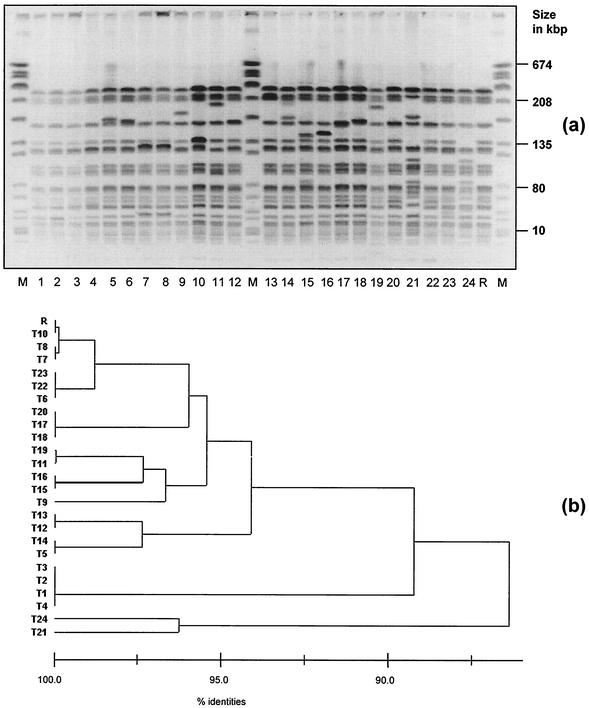
Macrorestriction patterns of all transconjugants were analyzed for clonal relatedness with the pattern of recipient 64/3 by PFGE (Fig. 1). Eleven of 24 transconjugants possessed a pattern 100% identical to that of the recipient, 11 transconjugants showed a pattern with only 1 to 3 band differences from the recipient's pattern and would be regarded as strongly related based on generally accepted recommendations (20, 21). Two transconjugants possessed a pattern with more than 3 band differences from the recipient and would be regarded as only widely related. However, the patterns of these two transconjugants, T21 and T24, were clearly different from the patterns of the donor isolates (data not shown).
FIG. 1.
(a) Macrorestriction patterns of SmaI-digested genomic DNA of 24 transconjugants and recipient R (negated gel image). Lanes: M, SmaI-digested genomic DNA of S. aureus NCTC 8325; 1 to 24, transconjugants 1 to 24; R, recipient isolate. (b) Cluster analysis according to the method of Claus et al. (8) by using a band-based similarity coefficient-generated matrix (Dice).
RAPD analysis.
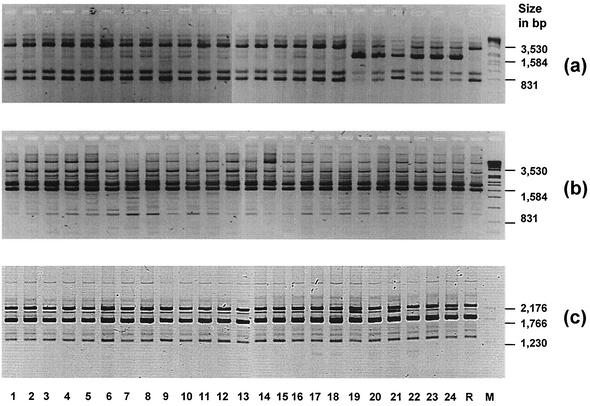
Different primers and conditions were applied for RAPD analysis of the transconjugants. Using primers and conditions described previously (16, 26), PCRs 2 and 3 did not discriminate between transconjugants and the recipient (Fig. 2b, c). PCR 1 clustered all transconjugants into two major groups: 18 transconjugants with a pattern identical to the recipient and 6 transconjugants with a pattern differing from the former ones by a single major fragment (Fig. 2a). The corresponding fragment in the pattern of transconjugant T19 was eluted from the agarose gel and purified via a commercial kit. The fragment was cloned into pUC18 and sequenced. The incorporated fragment possessed a complete open reading frame (GenBank accession number AY081910). It possessed 100% identity with tetL. The putative gene product of tetL confers resistance to all tetracycline antibiotics by efflux via a membrane-bound ABC porter. All 12 tetracycline-resistant transconjugants were tested by PCR for tetM and tetL. Five isolates, for which the MICs of oxytetracycline were 8 to 16 μg/ml, possessed only tetL, whereas the other seven isolates, for which the MICs of oxytetracycline were ≥256 μg/ml, possessed both determinants.
FIG. 2.
PCR RAPD typing for 24 transconjugants (lanes 1 to 24) and recipient R (negated gel image). (a) RAPD1 with primer 1; (b) RAPD2 with primer 2; (c) RAPD3 with primer 3. Lane M, molecular size standard. In panels a and b, SPP1 DNA was cleaved with EcoRI; panel c is a mix of pBR322 cleaved with Bgl2 and HinfI.
AFLP analysis.
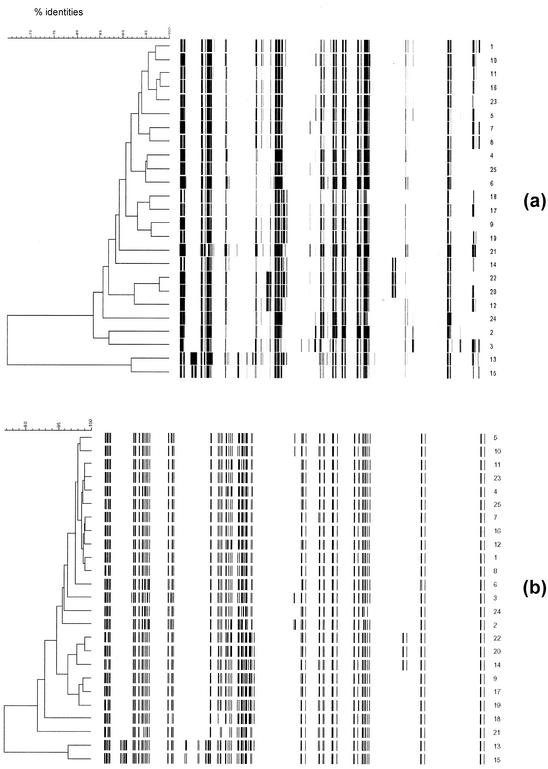
Analysis of the clustering of the AFLP patterns revealed that the level of similarities of AFLP banding patterns was strongly dependent on the coefficient used for calculating similarities, the curve-based Pearson correlation or the band-based Dice coefficient. The most common evaluation method used for AFLP is UPGMA clustering of the Pearson correlation. Based on repetitive analysis of identical samples, it is generally accepted that patterns with more than 90% identity indicate related isolates (33). Comparing the Pearson correlation similarity coefficient of the 24 transconjugants with that of the recipient showed that only 14 transconjugants exhibited AFLP patterns >90% similar (Fig. 3a). Eight isolates gave patterns with between 83 and 90% identity with the recipient, and two isolates possessed completely different patterns, with less than 70% similarity. Using a band-based similarity coefficient (Dice), the AFLP patterns of 22 transconjugants were >90% similar to that of the recipient (Fig. 3b). Only two isolates harbored patterns which did not belong to this group. Fragments in the pattern of recipient R appeared in all other patterns.
FIG. 3.
AFLP typing for 24 transconjugants (1 to 24) and recipient (25). (a) Pearson coefficient evaluation; (b) Dice coefficient evaluation.
Plasmid analysis.
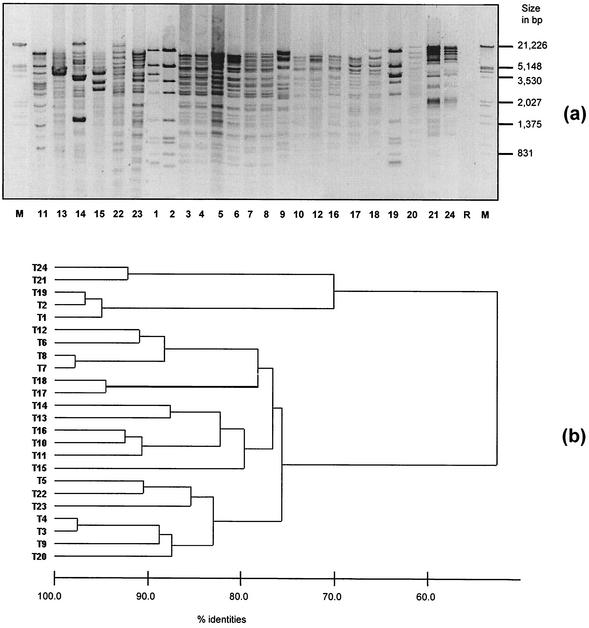
HindIII-digested plasmid DNA from all isolates showed only minor similarities after separation in an agarose gel (Fig. 4). However, in some transconjugants, identical or highly similar plasmid patterns were found, like in transconjugants 1, 2, and 19; 21 and 24; 7 and 8; 10 and 16; 3 and 4; and 17 and 18. Different antibiotic resistance determinants partly used as selective markers in the mating experiments have previously been demonstrated on the corresponding plasmids (28, 30).
FIG. 4.
(a) Patterns of HindIII-digested plasmids from transconjugants 1 to 24 and recipient R (negated gel image). M, Molecular size marker (lambda DNA cleaved with EcoRI and HindIII). (b) Cluster analysis according to the method of Claus et al. (8) by using a band-based similarity coefficient-generated matrix (Dice).
DISCUSSION
A variety of molecular methods, such as RAPD, PFGE, and AFLP, have successfully been applied to bacterial typing (15, 23, 24). In restriction-based techniques, such as PFGE and ribotyping, the appearance and disappearance of fragments can be postulated or modulated in theoretical and practical models (10, 13, 20, 21, 22). Based on this, a differentiation of major or minor relatedness between isolates can be clearly derived and is widely accepted by the scientific community. For amplification-based techniques (AFLP and RAPD), such criteria do not exist because amplification of distinct fragments is random and dependent on several amplification conditions (annealing temperature, PCR machine, polymerase, and polymerization time, etc.). However, these methods are also highly reliable when differences in experimental technology in inter- and intralaboratory tests have been kept to an achievable minimum (3, 26, 27). Compared with the gold standard, PFGE, the newer methods have demonstrated reproducible results with a high degree of discriminatory power when tested with already characterized natural strains. We chose a different approach in our study: minor, definite changes in the genome of our test strains were introduced via conjugative transfer of foreign DNA.
Fragment patterns of PFGE for 22 of the 24 transconjugants tested in this study were homogenous. Antibiotic resistance determinants in E. faecium have been recently demonstrated to be mostly plasmid-borne and can be used as selective markers in conjugation experiments (28, 30). Plasmids do not appear as fragments in PFGE except when they possess at least one or several recognition sites for SmaI and appear as linear fragment(s) (4). This would result in additional bands in the patterns of the transconjugants, with their number corresponding to the number of recognition sites for SmaI on the plasmids. As shown recently, these linearized plasmid bands can also appear to be the same size as chromosomal fragments from the recipient and are thus only detectable after Southern hybridization with a labeled probe (Werner et al., letter). Major fragment shifts only appeared in the PFGE patterns of two transconjugants, indicating that in most strains plasmid restriction fragments resulted in less than 3 band differences in the PFGE banding patterns of the transconjugants compared to the pattern of recipient R. Based on the suggested agreements, these 22 transconjugants would be regarded as strongly related and the other two would be regarded as related to the pattern of the recipient (20, 21).
Three different RAPD approaches were chosen for a typing of the 24 transconjugants and the recipient. Primers and RAPD assay conditions used in this study discriminated between the different donor isolates and the recipient strain (data not shown) (16, 26). For the transconjugants, two RAPD primer combinations did not allow any discrimination between the recipient and the 24 transconjugant strains. For the third primer set, RAPD1, fragment patterns for the transconjugants were either completely identical or differed by a major fragment that harbored the resistance gene tetL. According to the generated fragment patterns, 19 samples (18 transconjugants and the recipient) would be regarded as identical and 6 samples showed a clearly different pattern. Intriguingly, all 12 tetracycline-resistant transconjugants were positive for tetL. Only 6 transconjugants generated a different RAPD1 pattern, indicating that tetL was not in an identical genetic background in all 12 transconjugants (primer RAPD1 binds outside the coding region of tetL). Therefore, tetL itself did not influence the pattern generated with RAPD1 PCR. The occurrence of tetL in all tetracycline-resistant transconjugants confirms the results of earlier and recent studies showing a wide distribution of tetL in E. faecium (1, 5). Additionally, tetL seems to be more easily transferable than tetM, which is often found in Tn916-related elements integrated in the chromosome of enterococci (5).
The results of AFLP experiments led to two major conclusions: (i) UPGMA-generated clusters are strongly dependent on the method used for evaluation and (ii) AFLP patterns are not strongly influenced by plasmids, at least in E. faecium. Percent similarities were strongly dependent on the method used for evaluation (Fig. 3). According to the results with a number of repetitive determinations with identical strains, isolates possessing patterns with more than 90% identity are regarded as strongly related (33). In the study presented here, however, using the Pearson correlation coefficient, fragment patterns were up to 35% heterogeneous, with only 14 transconjugants and the recipient being more than 90% identical. Using the Dice coefficient, the evaluation of 22 transconjugants and the recipient showed more than 90% identity. The two transconjugants possessing more than 10% heterogeneity were not identical with the isolates showing more than a 3-band difference in PFGE. Based on these results, the criteria for genetic relatedness of isolates should be adjusted when similarities are calculated by using the Pearson correlation. Instead of using the 90% limit for describing closely related strains, we recommend using 80% similarity.
Recently, the host specificity of E. faecium in different animal, human, and environmental samples has been demonstrated with RAPD and AFLP, suggesting a wider epidemiological approach for these methods. For example, AFLP results indicated that isolates appeared to cluster according to their ecological origins (33). Isolates from infections in humans were clearly different from colonizers of the intestinal tracts of animals or humans. However, these isolates were mostly from The Netherlands, limiting the overall conclusions of this study. Additional studies with pig and poultry isolates from other countries were also clustered within their appropriate groups, or ecovars, and appeared to confirm the Dutch experience (6, 7, 33; M. Soltani, R. J. L. Willems, M. van Santen-Verheuvel, J. Philpott-Howard, D. Beighton, and N. Woodford, unpublished data). In a recent study, E. faecium typed with AFLP and RAPD revealed similar clusters of identity, but the isolates did not group according to their geographical or ecological origins (25). The major disadvantage of this later study was the small number of isolates from typical enterococcal infections analyzed compared to the number of food isolates (25). These contrasting findings for AFLP serve to emphasize the necessity for a standardized and proven methodology for using AFLP in E. faecium (25, 32, 33). Curve-based Pearson product moment correlation, which is the most common method used to calculate similarities, has major disadvantages. By using this method, signal strength and background signals, which are visible as nonspecific peaks, contribute to the measured similarity between AFLP patterns (Fig. 3a). A manual evaluation of assigning bands in AFLP patterns and calculating the alternative band-based Dice coefficient is more labor-intensive but produces more-reliable data, especially for strongly related isolates. This has to be kept in mind when using AFLP for epidemiological questions with E. faecium and other bacteria in general.
Digested plasmids showed heterogeneous patterns when resolved in agarose gels (Fig. 4). Most transconjugants with at least similar plasmid patterns also possessed similar PFGE and AFLP patterns (e.g., T7 and T8, T10 and T16, and T17 and T18), and as indicated here, plasmid fingerprints could be used as an additional typing tool for strongly related isolates of E. faecium. The efficacy of this approach has recently been demonstrated for strains with identical PFGE patterns isolated during an outbreak in a bone marrow transplantation unit in the United Kingdom (35) and for vancomycin-resistant Enterococcus strains from various hospitals in Germany (17).
To summarize, we compared E. faecium transconjugants differing by the possession of nonidentical resistance plasmids with typing methods commonly used for these bacteria. Without major reservation, all three methods could be used to type these strongly related bacteria. It was shown that, more than a decade after its introduction into molecular biology, PFGE macrorestriction patterns still could be recommended as the gold standard for studying local outbreaks. RAPD PCR typing cannot detect minor changes in the genome of the bacteria as introduced with our approach. In addition, with the already-known limitations of that methodology (comparably low reproducibility, low discriminatory power when compared to AFLP and PFGE), we would not recommend RAPD as a standard method for the replacement of PFGE or AFLP. However, under standardized conditions, it could be a quite useful and quick method. Fragment patterns generated with the AFLP approach obviously are not as strongly influenced by plasmid contents as sometimes discussed. Similarity matrices and resulting cluster analysis are very highly dependent on the method used for evaluation of the fragment patterns. Results of recent studies have shown that AFLP and RAPD might be used for epidemiological investigations of wider relatedness between enterococci and related gram-positive bacteria. Sequence information of complete genomes provide an excellent tool for an evaluation of the AFLP approach. Fragments appearing in AFLP patterns of corresponding strains become predictable and allow a fine tuning of the AFLP protocol (A. van Belkum, personal communication). For studying global epidemiology and tracking the worldwide interhospital spread of virulent, epidemic, and multiresistant clones and to provide an unambiguous nomenclature for E. faecium genotypes, multilocus sequence typing would be the method of choice. At least in cases of outbreaks with enterococci, plasmid profiles could be used as an additional tool when the need for better discrimination is beyond the capabilities of other typing methods.
Acknowledgments
The skillful technical assistance of J. Top and K. Guenther is highly acknowledged.
This work was supported by grant 325-4471-02/41 from the Federal Ministry for Health, Germany.
REFERENCES
- 1.Aarestrup, F. M., Y. Agerso, P. Gerner-Smidt, M. Madsen, and L. B. Jensen. 2000. Comparison of antimicrobial resistance phenotypes and resistance genes in Enterococcus faecalis and Enterococcus faecium from humans in the community, broilers, and pigs in Denmark. Diagn. Microbiol. Infect. Dis. 37:127-137. [DOI] [PubMed] [Google Scholar]
- 2.Antonishyn, N. A., R. R. McDonald, E. L. Chan, G. Horsman, C. E. Woodmansee, P. S. Falk, and C. G. Mayhall. 2000. Evaluation of fluorescence-based amplified fragment length polymorphism analysis for molecular typing in hospital epidemiology: comparison with pulsed-field gel electrophoresis for typing strains of vancomycin-resistant Enterococcus faecium. J. Clin. Microbiol. 38:4058-4065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barbier, N., P. Saulnier, E. Chachaty, S. Dumontier, and A. Andremont. 1996. Random amplified polymorphic DNA typing versus pulsed-field gel electrophoresis for epidemiological typing of vancomycin-resistant enterococci. J. Clin. Microbiol. 34:1096-1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barton, B. M., G. P. Harding, and A. Zuccharelli. 1995. A general method for detecting and sizing large plasmids. Anal. Biochem. 226:235-240. [DOI] [PubMed] [Google Scholar]
- 5.Bentorcha, F., G. de Cespédès, and T. Horaud. 1991. Tetracycline resistance heterogeneity in Enterococcus faecium. Antimicrob. Agents Chemother. 35:808-812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Borgen, K., Y. Wasteson, H. Kruse, and R. J. L. Willems. 2002. Vancomycin-resistant Enterococcus faecium (VREF) from Norwegian poultry cluster with VREF from poultry from the United Kingdom and The Netherlands in an amplified fragment length polymorphism genogroup. Appl. Environ. Microbiol. 68:3133-3137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bruinsma, N., R. J. L. Willems, A. E. van den Bogaard, M. van Santen-Verheuvel, N. London, C. Driessen, and E. E. Stobberingh. 2002. Different levels of genetic homogeneity in vancomycin-resistant and -susceptible Enterococcus faecium isolates from different human and animal sources analyzed by amplified-fragment length polymorphism. Antimicrob. Agents Chemother. 46:2779-2783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Claus, H., C. Cuny, B. Pasemann, and W. Witte. 1996. A database system for fragment patterns of genomic DNA of Staphylococcus aureus. Bundesgesundheitsblatt 2:68-74. [DOI] [PubMed] [Google Scholar]
- 9.Enright, M. C., and B. G. Spratt. 1999. Multilocus sequence typing. Trends Microbiol. 7:482-487. [DOI] [PubMed] [Google Scholar]
- 10.Goering, R. 1998. The molecular epidemiology of nosocomial infection-an overview of principles, application, and interpretation, p. 131-157. In S. Specter (ed.), Rapid detection of infectious agents. Plenum Press, New York, N.Y.
- 11.Homan, W. L., D. Tribe, S. Poznanski, M. Li, G. Hogg, E. Spalburg, J. D. A. van Embden, and R. J. L. Willems. 2002. A multilocus sequence typing scheme for Enterococcus faecium. J. Clin. Microbiol. 40:1963-1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Klare, I., E. Collatz, S. Al-Obeid, J. Wagner, A. C. Rodloff, and W. Witte. 1992. Glykopeptidresistenz bei Enterococcus faecium aus Besiedlungen und Infektionen von Patienten aus Intensivstationen Berliner Kliniken und einem Transplantationszentrum. ZAC Zschr. Antimikrob. Antineoplast. Chemother. 10:45-53. [Google Scholar]
- 13.Morrison, D., N. Woodford, P. Barrett, P. Sisson, and B. E. Cookson. 1999. DNA banding pattern polymorphism in vancomycin-resistant Enterococcus faecium and criteria for defining strains. J. Clin. Microbiol. 37:1084-1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Murray, B. E., K. V. Singh, J. D. Heath, B. R. Sharma, and G. M. Weinstock. 1990. Comparison of genomic DNAs of different enterococcal isolates using restriction endonucleases with infrequent cutting sites. J. Clin. Microbiol. 28:2059-2063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Olive, D. M., and P. Bean. 1999. Principles and applications of methods for DNA-based typing of microbial organisms. J. Clin. Microbiol. 37:1661-1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Quednau, M., S. Ahrne, A. C. Petersson, and G. Molin. 1998. Antibiotic-resistant strains of Enterococcus isolated from Swedish and Danish retailed chicken and pork. J. Appl. Microbiol. 84:1163-1170. [DOI] [PubMed] [Google Scholar]
- 17.Reinert, R. R., G. Conrads, J. J. Schlaeger, G. Werner, W. Witte, R. Lütticken, and I. Klare. 1999. Survey of antibiotic resistance among enterococci in North Rhine-Westphalia, Germany. J. Clin. Microbiol. 37:1638-1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 19.Savelkoul, P. H., H. J. Aarts, J. de Haas, L. Dijkshoorn, B. Duim, M. Otsen, J. L. Rademaker, L. Schouls, and J. A. Lenstra. 1999. Amplified-fragment length polymorphism analysis: the state of an art. J. Clin. Microbiol. 37:3083-3091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tenover, F. C., R. D. Arbeit, R. V. Goering, P. A. Mickelson, B. E. Murray, D. H. Persing, and B. Swaminathan. 1995. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J. Clin. Microbiol. 33:2233-2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tenover, F. C., R. D. Arbeit, and R. V. Goering. 1997. How to select and interpret molecular strain typing methods for epidemiological studies of bacterial infections: a review for healthcare epidemiologists. Molecular Typing Working Group of the Society for Healthcare Epidemiology of America. Infect. Control Hosp. Epidemiol. 18:426-439. [DOI] [PubMed] [Google Scholar]
- 22.Thal, L., J. Silverman, S. Donabedian, and M. Zervos. 1997. The effect of Tn916 insertions on contour-clamped homogeneous electrophoresis patterns of Enterococcus faecalis. J. Clin. Microbiol. 35:969-972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.van Belkum, A., P. W. M. Hermans, L. Licciardello, S. Stefani, W. Grubb, W. van Leeuwen, and W. H. F. Goessens. 1998. Polymerase chain reaction-mediated typing of microorganisms: tracking dissemination of genes and genomes. Electrophoresis 19:602-607. [DOI] [PubMed] [Google Scholar]
- 24.van Belkum, A. 1996. Molecular typing methods for epidemiological studies in medical microbiology. Med. Microbiol. Lett. 5:271-283. [Google Scholar]
- 25.Vancanneyt, M., A. Lombardi, C. Andrighetto, E. Knijff, S. Torriani, K. J. Bjorkroth, C. M. Franz, M. R. Foulquie Moreno, H. Revets, L. De Vuyst, J. Swings, K. Kersters, F. Dellaglio, and W. H. Holzapfel. 2002. Intraspecies genomic groups in Enterococcus faecium and their correlation with origin and pathogenicity. Appl. Environ. Microbiol. 68:1381-1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.van den Braak, N., E. Power, R. Anthony, H. P. Endtz, H. A. Verbrugh, and A. van Belkum. 2000. Random amplification of polymorphic DNA versus pulsed field gel electrophoresis of SmaI DNA macrorestriction fragments for typing strains of vancomycin-resistant enterococci. FEMS Microbiol. Lett. 192:45-52. [DOI] [PubMed] [Google Scholar]
- 27.van Leeuwen, W., M. Sijmons, J. Sluijs, H. Verbrugh, and A. van Belkum. 1996. On the nature and use of randomly amplified DNA from Staphylococcus aureus. J. Clin. Microbiol. 34:2770-2777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Werner, G., I. Klare, and W. Witte. 1997. Arrangement of the vanA gene cluster in enterococci of different ecological origin. FEMS Microbiol. Lett. 155:55-61. [DOI] [PubMed] [Google Scholar]
- 29.Werner, G., I. Klare, H. Heier, K.-H. Hinz, G. Böhme, M. Wendt, and W. Witte. 2000. Quinupristin/dalfopristin-resistant enterococci of the satA(vatD) and satG(vatE) genotypes from different ecological origins in Germany. Microb. Drug Resist. 6:37-47. [DOI] [PubMed] [Google Scholar]
- 30.Werner, G., B. Hildebrandt, I. Klare, and W. Witte. 2000. Linkage of determinants for streptogramin A, macrolide-lincosamide-streptogramin B, and chloramphenicol resistance on a plasmid in Enterococcus faecium and dissemination of this cluster among streptogramin-resistant enterococci. Int. J. Med. Microbiol. 290:543-548. [DOI] [PubMed] [Google Scholar]
- 31.Werner, G., B. Hildebrandt, and W. Witte. 2001. A cluster of aminoglycoside-streptothricin resistance genes aadE-sat4-aphA-3 disseminated among multiresistant isolates of Enterococcus faecium. Antimicrob. Agents Chemother. 45:3267-3269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Willems, R. J., W. Homan, J. Top, M. van Santen-Verheuvel, D. Tribe, X. Manzioros, C. Gaillard, C. M. Vandenbroucke-Grauls, E. M. Mascini, E. van Kregten, J. D. van Embden, and M. J. Bonten. 2001. Variant esp gene as a marker of a distinct genetic lineage of vancomycin-resistant Enterococcus faecium spreading in hospitals. Lancet 357:853-855. [DOI] [PubMed] [Google Scholar]
- 33.Willems, R. J. L., J. Top, N. van den Braak, A. van Belkum, H. P. Endtz, D. Mevius, E. E. Stobberingh, A. van den Bogaard, and J. D. A. van Embden. 2000. Host specificity of vancomycin-resistant Enterococcus faecium. J. Infect. Dis. 182:816-823. [DOI] [PubMed] [Google Scholar]
- 34.Woodford, N., D. Morrison, B. Cookson, and R. C. George. 1993. Comparison of high-level gentamicin-resistant Enterococcus faecium isolates from different continents. Antimicrob. Agents Chemother. 37:681-684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Woodford, N., P. R. Chadwick, D. Morrison, and B. D. Cookson. 1997. Strains of glycopeptide-resistant Enterococcus faecium can alter their van genotypes during an outbreak. J. Clin. Microbiol. 35:2966-2968. [DOI] [PMC free article] [PubMed] [Google Scholar]