Abstract
Numerous regulatory structural motifs have been identified as playing essential roles in transcriptional and post-transcriptional regulation of gene expression. RegRNA is an integrated web server for identifying the homologs of regulatory RNA motifs and elements against an input mRNA sequence. Both sequence homologs and structural homologs of regulatory RNA motifs can be recognized. The regulatory RNA motifs supported in RegRNA are categorized into several classes: (i) motifs in mRNA 5′-untranslated region (5′-UTR) and 3′-UTR; (ii) motifs involved in mRNA splicing; (iii) motifs involved in transcriptional regulation; (iv) riboswitches; (v) splicing donor/acceptor sites; (vi) inverted repeats; and (vii) miRNA target sites. The experimentally validated regulatory RNA motifs are extracted from literature survey and several regulatory RNA motif databases, such as UTRdb, TRANSFAC, alternative splicing database (ASD) and miRBase. A variety of computational programs are integrated for identifying the homologs of the regulatory RNA motifs. An intuitive user interface is designed to facilitate the comprehensive annotation of user-submitted mRNA sequences. The RegRNA web server is now available at http://RegRNA.mbc.NCTU.edu.tw/.
INTRODUCTION
A substantial number of mRNA structural motifs have been identified as playing essential roles in transcriptional and post-transcriptional regulation of gene expression, including transcription termination, mRNA localization and stability, mRNA alternative splicing and translation efficiency (1). For instance, riboswitches are complex folded RNA domains found in non-coding parts of various mRNAs, where they control gene expression by harnessing allosteric structural changes (2). In numerous cases, specific functional RNA elements whose biological activity relies on their primary sequences and specific secondary structures, act either as target sites for RNA-binding factors or directly interact with translation machinery (3). Since ligand-binding and base pairing are fundamental features of RNA interactions and activities, many of such small regulatory RNA motifs likely exist in cells (4).
Previous research indicates that exonic splicing enhancer (ESE) is a binding site of serine/arginine-rich proteins (SR proteins), which belong to a family of conserved splicing factors and were first implicated in splicing when discovered that they are components of the spliceosome (5). MicroRNAs (miRNAs) are small RNA molecules, which are ∼22 nt sequences play critical roles in translational regulation and degradation of mRNA by hybridizing to miRNA target sites within the 3′-untranslated region (3′-UTR) of the mRNAs.
Rfam is a comprehensive collection of non-coding RNA (ncRNA) families, represented by multiple sequence alignments and profile stochastic context-free grammars (6). It facilitates the identification and classification of new members of known sequence families, and distributes annotation of ncRNAs in over 200 complete genome sequences.
The PatSearch software (7) can search user-submitted sequences for any combination of sequence patterns represented as positional weighted matrix (PWMs) and structural motifs and allow the mismatch and mispairing search under a user-designated threshold. Transterm (8) provides data for principal regions, such as UTRs and ORFs, in mRNA sequences and regulatory RNA motifs, and allows users to investigate their own motifs or mRNA sequences. These motifs are typically located in the 3′-UTR, 5′-UTR and coding regions. Riboswitch finder (9) and RibEx (10) were developed to determine known riboswitches in a given sequence. UTRscan (11) allows users searching in submitted sequences for regulatory motifs collected in UTRsite (3). ESEfinder (12) is a web resource for identifying a series of ESEs in mRNA sequences. RNAMotif (13) devises the definition of RNA structural descriptor to describe an RNA structure motif and scan the input sequences for structural homologs of regulatory RNA motifs and elements (13).
Survey results indicate that various approaches can be used for predicting regulatory RNA motifs. Additionally, various databases and web servers have accumulated regulatory RNA motifs during the last few decades. Therefore, in order to annotate regulatory RNA motifs in messenger RNAs, the integration of numerous databases of regulatory RNA motifs and computational analytical tools are crucial.
The purpose of this work is to develop an integrated web server, RegRNA, to identify homologs of the regulatory RNA motifs and elements against an input mRNA sequence. Both sequence homologs and structural homologs of regulatory RNA motifs can be recognized. RegRNA not only extracts known regulatory RNA motifs by surveying literatures and several regulatory RNA motif databases, such as UTRdb (3), TRANSFAC (14), alternative splicing database (ASD) (15) and miRBase (16), but also collects known regulatory RNA motifs including motifs in 5′-UTR and 3′-UTR, and motifs involved in transcriptional regulation, exonic/intronic splicing motifs, splicing donor/acceptor sites, inverted repeats, and miRNA target sites. A variety of computational programs for different types of regulatory RNA motifs were also implemented. Moreover, RegRNA displays prediction results in a graphical interface generated by various integrated analytical tools, and allows users to annotate their own experimental sequences or to discover homologs of their desired motifs.
DESIGN AND IMPLEMENTATION
Data collection
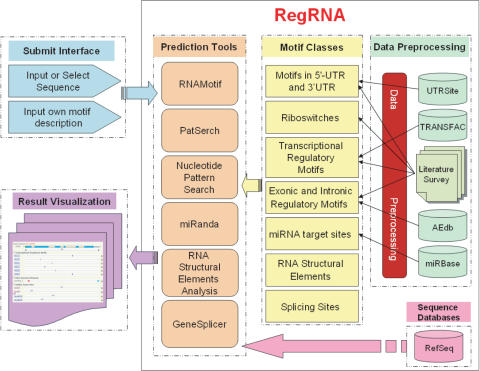
The regulatory RNA motifs supported in RegRNA are categorized into several classes: (i) motifs in mRNA 5′-UTR and 3′-UTR; (ii) motifs involved in mRNA splicing; (iii) motifs involved in transcriptional regulation; (iv) riboswitches; (v) splicing donor/acceptor sites; (vi) inverted repeat; and (vii) miRNA target sites. Figure 1 shows the information and processing flow of RegRNA
Figure 1.
RegRNA information and processing flow.
This work collected known regulatory RNA motifs and elements via literature survey and by integrating a variety of regulatory RNA motif databases, such as UTRSite (3), TRANSFAC (14), ASD (15), miRBase (16) and miRNAMap (17). The UTRSite (Release 16) stores a collection of regulatory RNA located in 5′-UTR and 3′-UTR, whose function and structure were experimentally determined (3). The TRANSFAC database (Release 7.4) (14) collects known transcriptional factors, their binding sites, nucleotide distribution matrices and regulated genes. Those transcriptional regulatory sites located in pre-mRNA or mature mRNA sequences were extracted. Such sites can occur in 5′-UTR, introns or exons. Riboswitches are natural genetic control elements typically located in UTR of mRNA sequence to form a binding pocket for a metabolite that regulates that gene expression. Other features, such as riboswitch structural motifs, were also extracted by surveying literatures.
The ASD is a literature-based dataset containing sequences and properties of alternatively spliced exons, functional enumeration of observed splicing events, and characterization of splicing regulatory elements (15). In addition to the well-known splice sites, such as donor sites and acceptor sites, the conserved sequences in exonic and intronic regions are reportedly involved in an alternative splicing mechanism. Besides the splicing motifs from ASD, numerous ESE and exonic splicing silencers (ESS) were obtained from literatures. The miRBase database (Release 7.1) (16) provides comprehensive microRNA sequence data, annotation and predicted gene targets. The known miRNA genes in three mammalian genomes—human, mouse and rat—were obtained from miRBase (16).
Therefore, the RegRNA currently collects 1274 regulatory motifs (Table 1). For instance, there are 40 regulatory RNA motifs located in 5′-UTR and 3′-UTR. The number of motifs involved in splicing is 43. Totally, 744 known miRNAs in humans, mice and rats were obtained.
Table 1.
Statistics of types of regulatory RNA motifs in RegRNA
| Types of regulatory RNA motifs | Number of entries | Data sources |
|---|---|---|
| In mRNA 5′-UTR and 3′-UTR | 40 | UTR site (3) |
| Riboswitches | 14 | Literature survey |
| Involved in splicing (exonic) | 176 | Literature survey, ASD (15) |
| Involved in splicing (intronic) | 91 | Literature survey, ASD (15) |
| Transcriptional regulation (exonic) | 21 | Literature survey, TRANSFAC (14) |
| Transcriptional regulation (intronic) | 156 | Literature survey, TRANSFAC (14) |
| Transcriptional regulation (UTR) | 22 | Literature survey, TRANSFAC (14) |
| miRNA (human, mouse and rat) | 744 | miRBase (16) |
| Total | 1274 |
Integrated analytical tools
As to the regulatory RNA motifs in the form of primary structures represented as consensus patterns or sequence patterns, RegRNA integrates several motif identification tools, such as EMBOSS—fuzznuc (18), EMBOSS—einverted (18), and GeneSplicer (19) to detect the homologs of the regulatory RNA motifs (Table 2).
Table 2.
Computational analysis tools applied in RegRNA
| Supported analyzing functions | Characteristics of regulatory motifs | Types of regulatory RNA motifs | Integrated tools |
|---|---|---|---|
| Consensus pattern search | Sequence pattern | Motifs involved in mRNA splicing and regulation of transcription | EMBOSS—fuzznuc (18) |
| Splicing sites | Sequence pattern | Prediction of the splice sites | GeneSplicer (19) |
| RNA structural elements | Secondary structure | Inverted repeat | EMBOSS—einverted (18) |
| RNA structural motif search | Secondary structure | Motifs in mRNA 5′-UTR, 3′-UTR and open reading frames | RNAMotif (13) |
| miRNA target sites | Duplex of nucleotide | miRNA target sites | miRanda (20) |
The fuzznuc of EMBOSS package is applied to search a consensus pattern against a sequence. The einverted of EMBOSS package is utilized to detect inverted repeats in a nucleotide sequence. GeneSplicer (19) was developed for determining splice sites in eukaryotic mRNA by combining several schemes that have already proven successful in characterizing the patterns surrounding donor and acceptor sites. Moreover, in identifying miRNA targets, miRanda (20) is employed to detect the miRNA target sites in a sequence that complimentary hybridizes to the mature miRNAs. The minimum free energy (MFE) of miRNA–target duplex is determined by miRanda when predicting miRNA target sites.
As to the regulatory RNA motifs in the form of secondary structures represented as RNA structural descriptors (13), RegRNA integrates RNAMotif to identify homologs of regulatory RNA structural motifs in user-submitted sequences (Table 2). Some of regulatory RNA motifs in 5′-UTR and 3′-UTR and riboswitches are represented as RNA structural descriptors in advance for searching the homologs against the user-submitted sequences.
INTERFACE
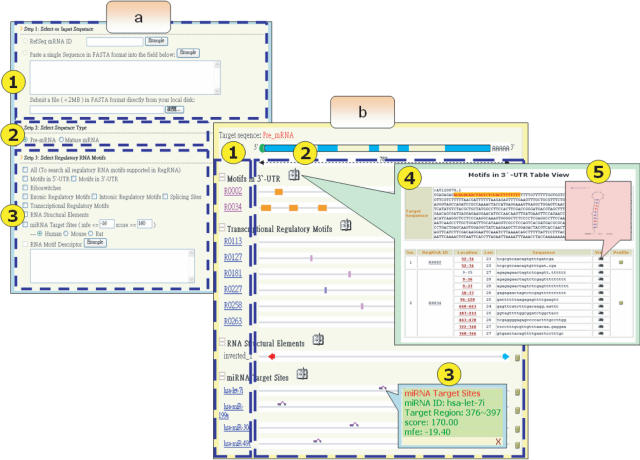
The RegRNA web server devised a variety of interface to facilitate the analysis for the homologs of the regulatory RNA motifs. Users can submit a sequence by inputting the accession number for retrieving mRNA sequence from RefSeq database (21), inputting a single sequence in FASTA format, or uploading a sequence file (Figure 2a, label 1). Subsequently, users select the sequence type—pre-mRNA or mature mRNA (Figure 2a, label 2). When the input sequence is specified as a mature mRNA sequence, the prediction of the splicing site and the identification of intronic regulatory RNA motifs are not applied. Finally, users must decide which types of regulatory RNA motifs to be investigated by just clicking the checkbox (Figure 2a, label 3). Additionally, in predicting miRNA target sites, users can adjust the MFE threshold and score threshold to filter miRNA targets of interest.
Figure 2.
(a) Prediction page of the RegRNA website. (b) The result web page to graphically reveal the location of the homologs in regulatory RNA motifs and elements.
Upon submission to the website, the input sequence is then analyzed and the prediction results are presented via both textual interface and graphical interface (Figure 2b). Users can dsitinguish clearly between different regulatory RNA motifs and link to detailed descriptions via hyperlinks. If the detected homologs are regulatory RNA structural motifs, the RNA secondary structure of the detected homologs are generated by mfold (22) and provided on the web.
CASE STUDY
In order to demonstrate the functionality of RegRNA, the authors use a case study, such as iron response element (IRE), which is a particular hairpin structure located in the 5′-UTR or 3′-UTR of various mRNAs involved in cellular iron metabolism (23). Supplementary Figure S1 (See Supplementary Data) illustrates how the IRE structural motif regulates iron concentrations. For example, two different IREs present their conservation of sequence and structure determinants (Supplementary Figure S2) (23).
Subsequently, RegRNA detected homologs of IRE motif in a given RNA sequence. For instance, a sequence with the accession number of EMBL, AF068225, was obtained from Rfam (6). Supplementary Figure S3 (See Supplementary Data) presents the predictive results of this case study. The IRE motif in the given RNA sequence was accurately identified by the proposed RegRNA.
DISCUSSION AND CONCLUSION
The RegRNA is capable of identifying homologs of plentiful types of regulatory RNA motifs. Table 3 gives the comparison of RegRNA with other previously developed tools including RNAMotif (13), PatSearch (7), RibEx (10), Riboswitch Finder (9), UTRScan (3) and Transterm (8). RibEx and Riboswitch Finder are developed to specially identify known riboswitches. Besides RNAMotif, all of the tools collected specific regulatory RNA motifs for identifying their homologs in sequences. RNAMotif, PatSearch and RegRNA allow the user-defined RNA structural motif for the analysis. In summary, the RegRNA provides more plentiful types of regulatory RNA motifs than other tools.
Table 3.
Comparison of RegRNA to other tools and web sites
| Comparing tools | Database support | RNA structural motif search | Consensus pattern search | Splicing sites | Inverted repeat | miRNA targets | User interface |
|---|---|---|---|---|---|---|---|
| RNA motif (13) | — | User-defined | Yes | — | — | — | — |
| PatSearch (7) | Yes | User-defined | Yes | — | — | — | Yes |
| RibEx (10) | Yes | Riboswitch | — | — | — | — | Yes |
| Riboswitch finder (9) | Yes | Riboswitch | — | — | — | — | Yes |
| UTR scan (11) | Yes | Motifs in 5′-UTR and 3′-UTR | Yes | — | — | — | Yes |
| Transterm (8) | Yes | Motifs in 5′-UTR, 3′-UTR and ORF | Yes | — | — | — | Yes |
| RegRNA | Yes | User-defined, Riboswitch, Motifs in 5′-UTR, 3′-UTR and ORF | Yes | Yes | Yes | Yes | Yes |
During the miRNA target prediction, the lower MFE values of the miRNAs and the target sites reveal that the energetically more probable hybridizations between the miRNAs and the target genes. Additionally, the predictive parameters including miRanda MFE and miRanda score were adjusted for the miRNA target prediction by comparing the predictive results to known miRNA/targets data according to our previous works (17). The MFE threshold of the miRNA and target duplex was suggested as −16 kcal/mol and the miRanda score was specified as 160.
Comparing the proposed RegRNA with Rfam database (6), the substantial difference between the two resources is that RegRNA aims on the annotation of regulatory RNA motifs and structural elements potentially involving in gene post-transcriptional regulation, while Rfam mainly aims on the collection of ncRNA families and a variety of regulatory RNA structural motifs. The authors compare the Rfam database and RegRNA web server in two aspects, the types of RNA data collected and the analyzing functions supported. Firstly, the comparing lists of the types of RNA data collected in the two resources are given in Supplementary Table S1 (See Supplementary Data). Rfam collects the ncRNA families including rRNA, tRNA, frameshift elements, riboswitches, thermoregulators, IRES and functional elements in UTR. However, RegRNA does not collect the ncRNA sequences, frameshift elements, thermoregulators and IRES. For the investigation the transcriptional and post-transcriptional regulation, RegRNA specially collects other regulatory RNA motifs including transcriptional regulatory motifs, splicing sites, exonic and intronic splicing motifs and microRNAs.
In the aspect of the supported analyzing functions (Supplementary Table S2, See Supplementary Data), RegRNA provides the annotation of the input RNA sequences for candidates of regulatory RNA structural motifs and regulatory RNA elements, which potentially involve in gene transcriptional and post-transcriptional regulation. Rfam facilitates the identification of the particular regions within the input sequences that are homologous to the ncRNAs and regulatory RNA motifs collected in Rfam. Moreover, our proposed RegRNA specially provides the identification of splicing sites, the detection of splicing-related regulatory motifs, the detection of transcriptional regulatory motifs, the detection of inverted repeats, and the identification of miRNA target sites in the input messenger RNA sequences.
Besides the regulatory RNA motifs collected in RegRNA, additional regulatory motifs were discovered in mRNA sequences, for example, pseudoknot, can be integrated into the RegRNA to make RegRNA more comprehensive. Pseudoknots occur in the secondary structure of numerous RNA molecules, such as ribosomal RNAs, the catalytic core of group I introns, RNase P RNAs and viral RNAs. In numerous cases, pseudoknots have functional roles in, e.g. ribosomal frameshifting, regulation of translation and splicing, and selinocystein biosynthesis (24). When considering to known miRNAs, miRNAMap (17) also collects putative miRNAs in human, mouse, rat and dog genomes. The putative miRNAs will be integrated into RegRNA in the future.
Although the proposed RegRNA provides comprehensive analysis for regulatory RNA motifs, it requires about 20 s if all kinds of the regulatory RNA motifs are selected for analysis. The conserved regions of the ncRNA families and the regulatory RNA motifs collected in Rfam are suitable for the RNA structural analyses supported in RegRNA. The authors plan to convert the consensus structures of ncRNA families in Rfam into the descriptors of RNA structural motifs, which can be maintained in RegRNA to support the annotation of regulatory RNA motifs against the input of RNA sequences. Consequently, reduction of the prediction time is another task. Perhaps, utilizing the concept of a distributed system to parallel predict different regulatory motifs will decrease the run time.
The RegRNA is an integrated web server for identifying known regulatory RNA motif in RNA sequences. The main contributions of this work are as follows: (i) the collection of the known regulatory RNA motifs and elements from literatures and multiple biological databases; (ii) the integration of a variety of motif identification tools for regulatory RNA motifs both in primary and secondary structure; (iii) providing intuitive interface to facilitate the presentation of plentiful information provided in the proposed RegRNA; (iv) providing a convenient solution for comprehensively annotating the mRNA sequence for regulatory mechanism involved by regulatory RNA motifs and elements.
AVAILABILITY
The RegRNA web server will be continuously maintained and updated. The web server is now freely available at http://RegRNA.mbc.nctu.edu.tw/.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
Acknowledgments
The authors would like to thank the National Science Council of the Republic of China for financially supporting this research under Contract No. NSC 95-3112-E-009-002. Special thanks for the financial support from the National Research Program For Genomic Medicine (NRPGM), Taiwan. This work was also partially supported by MOE ATU. Funding to pay the Open Access publication charges for this article was provided by National Science Council of the Republic of China.
Conflict of interest statement. None declared.
REFERENCES
- 1.Ji Y., Xu X., Stormo G.D. A graph theoretical approach for predicting common RNA secondary structure motifs including pseudoknots in unaligned sequences. Bioinformatics. 2004;20:1591–1602. doi: 10.1093/bioinformatics/bth131. [DOI] [PubMed] [Google Scholar]
- 2.Mandal M., Breaker R.R. Gene regulation by riboswitches. Nature Rev. Mol. Cell. Biol. 2004;5:451–463. doi: 10.1038/nrm1403. [DOI] [PubMed] [Google Scholar]
- 3.Mignone F., Grillo G., Licciulli F., Iacono M., Liuni S., Kersey P.J., Duarte J., Saccone C., Pesole G. UTRdb and UTRsite: a collection of sequences and regulatory motifs of the untranslated regions of eukaryotic mRNAs. Nucleic Acids Res. 2005;33:D141–D146. doi: 10.1093/nar/gki021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Laserson U., Gan H.H., Schlick T. Predicting candidate genomic sequences that correspond to synthetic functional RNA motifs. Nucleic Acids Res. 2005;33:6057–6069. doi: 10.1093/nar/gki911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cartegni L., Chew S.L., Krainer A.R. Listening to silence and understanding nonsense: exonic mutations that affect splicing. Nature Rev. Genet. 2002;3:285–298. doi: 10.1038/nrg775. [DOI] [PubMed] [Google Scholar]
- 6.Griffiths-Jones S., Moxon S., Marshall M., Khanna A., Eddy S.R., Bateman A. Rfam: annotating non-coding RNAs in complete genomes. Nucleic Acids Res. 2005;33:D121–D124. doi: 10.1093/nar/gki081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Grillo G., Licciulli F., Liuni S., Sbisa E., Pesole G. PatSearch: a program for the detection of patterns and structural motifs in nucleotide sequences. Nucleic Acids Res. 2003;31:3608–3612. doi: 10.1093/nar/gkg548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jacobs G.H., Stockwell P.A., Tate W.P., Brown C.M. Transterm--extended search facilities and improved integration with other databases. Nucleic Acids Res. 2006;34:D37–D40. doi: 10.1093/nar/gkj159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bengert P., Dandekar T. Riboswitch finder–a tool for identification of riboswitch RNAs. Nucleic Acids Res. 2004;32:W154–W159. doi: 10.1093/nar/gkh352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Abreu-Goodger C., Merino E. RibEx: a web server for locating riboswitches and other conserved bacterial regulatory elements. Nucleic Acids Res. 2005;33:W690–W692. doi: 10.1093/nar/gki445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pesole G., Liuni S. Internet resources for the functional analysis of 5′ and 3′ untranslated regions of eukaryotic mRNAs. Trends Genet. 1999;15:378. doi: 10.1016/s0168-9525(99)01795-3. [DOI] [PubMed] [Google Scholar]
- 12.Cartegni L., Wang J., Zhu Z., Zhang M.Q., Krainer A.R. ESEfinder: a web resource to identify exonic splicing enhancers. Nucleic Acids Res. 2003;31:3568–3571. doi: 10.1093/nar/gkg616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Macke T.J., Ecker D.J., Gutell R.R., Gautheret D., Case D.A., Sampath R. RNAMotif, an RNA secondary structure definition and search algorithm. Nucleic Acids Res. 2001;29:4724–4735. doi: 10.1093/nar/29.22.4724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Matys V., Kel-Margoulis O.V., Fricke E., Liebich I., Land S., Barre-Dirrie A., Reuter I., Chekmenev D., Krull M., Hornischer K., et al. TRANSFAC and its module TRANSCompel: transcriptional gene regulation in eukaryotes. Nucleic Acids Res. 2006;34:D108–D110. doi: 10.1093/nar/gkj143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stamm S., Riethoven J.J., Le Texier V., Gopalakrishnan C., Kumanduri V., Tang Y., Barbosa-Morais N.L., Thanaraj T.A. ASD: a bioinformatics resource on alternative splicing. Nucleic Acids Res. 2006;34:D46–D55. doi: 10.1093/nar/gkj031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Griffiths-Jones S., Grocock R.J., van Dongen S., Bateman A., Enright A.J. miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res. 2006;34:D140–D144. doi: 10.1093/nar/gkj112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hsu P.W., Huang H.D., Hsu S.D., Lin L.Z., Tsou A.P., Tseng C.P., Stadler P.F., Washietl S., Hofacker I.L. miRNAMap: genomic maps of microRNA genes and their target genes in mammalian genomes. Nucleic Acids Res. 2006;34:D135–D139. doi: 10.1093/nar/gkj135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rice P., Longden I., Bleasby A. EMBOSS: the European Molecular Biology Open Software Suite. Trends Genet. 2000;16:276–277. doi: 10.1016/s0168-9525(00)02024-2. [DOI] [PubMed] [Google Scholar]
- 19.Pertea M., Lin X., Salzberg S.L. GeneSplicer: a new computational method for splice site prediction. Nucleic Acids Res. 2001;29:1185–1190. doi: 10.1093/nar/29.5.1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Enright A.J., John B., Gaul U., Tuschl T., Sander C., Marks D.S. MicroRNA targets in Drosophila. Genome Biol. 2003;5:R1. doi: 10.1186/gb-2003-5-1-r1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wheeler D.L., Barrett T., Benson D.A., Bryant S.H., Canese K., Chetvernin V., Church D.M., DiCuccio M., Edgar R., Federhen S., et al. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2006;34:D173–D180. doi: 10.1093/nar/gkj158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hentze M.W., Kuhn L.C. Molecular control of vertebrate iron metabolism: mRNA-based regulatory circuits operated by iron, nitric oxide, and oxidative stress. Proc. Natl Acad. Sci. USA. 1996;93:8175–8182. doi: 10.1073/pnas.93.16.8175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ren J., Rastegari B., Condon A., Hoos H.H. HotKnots: heuristic prediction of RNA secondary structures including pseudoknots. RNA. 2005;11:1494–1504. doi: 10.1261/rna.7284905. [DOI] [PMC free article] [PubMed] [Google Scholar]