Abstract
KinDOCK is a new web server for the analysis of ATP-binding sites of protein kinases. This characterization is based on the docking of ligands already co-crystallized with other protein kinases. A structural library of protein kinase–ligand complexes has been extracted from the Protein Data Bank (PDB). This library can provide both potential ligands and their putative binding orientation for a given protein kinase. After protein–protein structural superposition, the ligands are transferred from the template complexes to the target protein kinase. The resulting complexes are evaluated using the program SCORE to compute a theoretical affinity. They can be dynamically visualized to allow a rapid mapping of important steric clashes and potential substitutions relevant for specificity and affinity. These characteristics allow a quick characterization of protein kinase active sites including conformation changes potentially required to accommodate particular ligands. Additionally, promising pharmacophores can be identified in the focussed library. These features will help to rationalize or optimize virtual screening (VS) on larger chemical compound libraries. The server and its documentation are freely available at http://abcis.cbs.cnrs.fr/kindock/.
INTRODUCTION
Structure-based methods are becoming increasingly important in drug discovery. This is especially true in the case of protein kinases, which are important therapeutic targets (1). While targeting the ATP-binding site of protein kinases was initially thought to be useless due to a lack of specificity, the first crystal structure of a protein kinase in 1991 revealed unexpected structural features leading to brighter hopes for the development of specific inhibitors (2). Since then, a number of protein kinase structures have been solved, including many inhibitor-bound proteins. A number of protein kinase inhibitors are now in the clinic and several others in clinical trials (1). Despite this progress, the gap between the number of reported sequences and experimental structures continues to increase (3), and we are far from determining the structure of all the 518 protein kinases identified in the human genome. Furthermore, a larger gap exists in the case of proteins complexed with their known ligands. At the same time, recurrent substructures are observed among the known ligands of a given protein family (or superfamily) as observed in the case of protein kinases (4). This observation, however has yet to be sufficiently exploited in docking by similarity.
Structure comparisons have been widely used to identify protein similarities and to derive functional or structural information (3). Automatic procedures are now available to provide three-dimensional models of good quality when the sequence identity is above 30% (5). On the contrary, the modelling of protein–ligand complexes remains a more difficult and tedious task (5). Indeed, template searches generally focus on sequence similarities rather than on the presence or not of a ligand bound to the template. In the case of well-characterized protein families, one template may provide the best protein scaffold while a more distantly related template may provide a valid ligand (e.g. ATP for protein kinases).
In parallel, virtual screening (VS) has successfully identified small chemical compounds showing micromolar to nanomolar affinities (6). While the use of VS had often been restricted to the structures of crystallized proteins, it was recently shown that some three-dimensional models can be accurate enough to perform VS (7,8). However, VS is still a highly CPU-intensive task and it needs an appropriate active site structure (e.g. corresponding to an active conformation). This conformation might differ from one class of compounds to another due to the ligand-induced fit phenomenon (8).
On the one hand, current comparative modelling can provide theoretical models of protein structures with significant accuracy. On the other hand, VS has been developed to search rather large chemical libraries and to identify potential ligands. Bridging both fields with new chemoinformatic tools could provide new approaches to drug design. However, because of a lack of appropriate tools (e.g. for local evaluation of protein conformations), the interplay between macromolecular modelling and ligand docking is not yet efficiently and automatically performed.
We present here a new server, kinDOCK, which has been developed to speed up these processes. It combines structural comparisons, immediate transfer of known ligands from the template structure into the target structure, visualization of the deduced protein–ligand complexes and evaluation of protein–ligand interactions. Using this server, the quality of a modelled structure (especially the active site or the ligand binding sites) as well as the conformation of a protein (active/open conformation, inactive/closed conformation) can be rapidly and precisely evaluated. It is also possible to identify protein templates in complex with a given ligand in order to restrain the active site modelling and take into account potential ligand induced-fit. Generic ligands or putative inhibitors can also be searched in our focused library prior to in vitro assays. Because protein kinases are important therapeutic targets, this server has been dedicated to the study of this class of enzymes (8,9). Several examples described herein, illustrate the various uses of kinDOCK in the new field of comparative docking.
METHODS
First, a set of protein kinase structures solved in complex with a ligand (e.g. ATP, inhibitors) was manually gathered from the Protein Data Bank (PDB) (http://www.rcsb.org/pdb); (10) and organized in a hierarchical database. A multiple-sequence alignment of these kinases based on the Hanks alignment (11) was manually refined using the alignment editor ViTO (12). A hidden Markov model (HMM) profile was then derived from this alignment. This profile can now be used to search automatically the PDB for protein kinase complexes to update our library. The bound protein kinase ligands were extracted and rewritten in mol2 format using the program PRODRG (http://davapc1.bioch.dundee.ac.uk/programs/prodrg/); (13).
The kinDOCK server needs, as an input data, either a coordinate file in PDB format or the four-letter PDB code of the target structure. The sequence of the protein kinase structure submitted to the server is aligned to the kinase profile using the program suite HMMER (http://hmmer.wustl.edu). Then, the target structure is superposed on each of the protein kinase complexes of our library according to the alignment. The superposition is based on a subset of Cα carbons defining the common binding site. The latter is composed of any aligned amino acid that possesses an atom at less than 6 Å from the ligand in the experimental complex. Then the ligands are transferred into the target binding site. The binding affinities of the new theoretical complexes are evaluated using the program SCORE (14). The affinity score (pK) predicted by SCORE (14) is −log(IC50) so the higher the score, the stronger the predicted affinity. Beside the predicted affinity scores, the sequence identity and the root mean square deviation (RMSD) of the active site superposition are displayed. Ranking the complexes according to these values or according to the ligand or structure PDB codes is also possible. The resulting complexes can be visualized directly using a java applet, Jmol (http://jmol.sourceforge.net, see Figure 1). Distances and angles can also be computed and visualized in the Jmol applet. The coordinates of the new complexes can be downloaded in PDB format for further analysis as well as the re-oriented ligands in MOL2 format for further analysis and virtual screening.
Figure 1.
Example of kinDOCK output. The cAMP-dependent protein kinase (PDB1STC) has been submitted to kinDOCK. The results were sorted by ligand name to highlight the scores and a binding mode obtained for staurosporine.
Features and case studies
KinDOCK allows a rapid and straightforward building of complexes using a three-dimensional structure (which may be experimental or theoretical) of a given protein kinase and a specialized library of protein kinase complexes. This structural library contains 310 crystal structures of protein kinase–ligand complexes representing currently 52 different protein kinases and 196 different ligands despite some redundancies for several compounds, such as ATP and its analogs and staurosporine. Automatic updates will be set up to follow the increasing number of complex structures solved.
KinDOCK performs the four following tasks: (i) the alignment of the sequence of the submitted protein kinase onto the family profile, (ii) the superposition of its three-dimensional structure on the protein kinase–ligands structures (according to the alignment), (iii) the extraction of the ligand from each complex and its transfer into the active site of the submitted structure and then (iv) the evaluation of the protein–ligand interactions. Herein, we called this approach ‘comparative docking’ by analogy with comparative modelling of protein structure. Currently, one comparative docking on the protein kinase family lasts a few minutes.
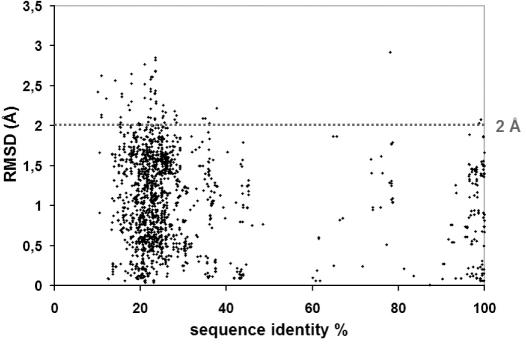
The orientation of a given ligand has been shown to be conserved among various protein kinases, especially for ligands containing at least three cycles (4). Thanks to protein–protein superposition, kinDOCK brings each ligand into the target active site with an orientation similar to the one found in the original ligand-kinase crystal structure. The deduced binding mode was expected to be more correct than one deduced by classical docking (15). Visual surveys suggested that the ligand orientation in the complexes deduced by kinDOCK closely resembles the experimental one. Figure 2 illustrates the case of staurosporine docked into PDB1STC from the 15 other complexes of a protein kinase with this generic inhibitor. Thanks to the redundancy of the complex library, an extensive evaluation of the quality of the molecule orientation predicted by kinDOCK was performed. For any compound found in at least two structures, the RMSD between the molecule posed by kinDOCK and the one in the PDB structure was computed. Most poses deduced by kinDOCK are within 2 Å from the experimental one (heavy atom RMSD; Figure 3) meaning that they are correct according to the standard threshold used in virtual screening. Furthermore, this appeared to be true whatever the sequence identity shared by the proteins (down to 20%).
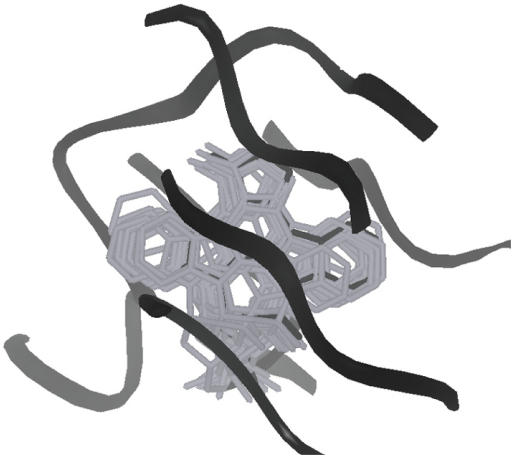
Figure 2.
Example of superposition by kinDOCK. All stauroporines—protein kinase complexes present in the PDB (16 in the current database; listed in Figure 1) were used to transfer the common ligand into the cAMP-dependent protein kinase (PDB1STC). The active site of cAMP-dependent protein kinase is drawn as a black ribbon. The experimental orientation of staurosporine (PDB1STC) is in black wireframe while the staurosporines docked by similarity are in grey wireframe.
Figure 3.
RMSD between the ligand poses as deduced by kinDOCK and the ligand in the template PDB structure.
The java viewer, Jmol, allows a rapid visualization of the protein–ligand contacts and a rapid survey of strong steric clashes and favourable interactions (salt bridge, ring stacking,…; see Figure 1). Alternatively, kinDOCK may also reveal an inaccurate modelling of the active site or a conformational rearrangement preventing the ligand binding.
Selection of active site conformations
As the set of complexes includes ATP-bound structures (and also ATP analog complexes) and other known binders (e.g. staurosporine, a generic inhibitor of protein kinases), the structure of the ATP binding site of a protein kinase can be probed. Indeed, kinDOCK can discriminate active and inactive forms of the same protein kinase. This was particularly evident for the crystal structures of the protein kinase PKB. KinDOCK was used to place ATP into the crystal structures of PKB, either in its active conformation (PDB1O6L) or in its inactive conformation (PDB1GZN). Protein–ligand interactions were evaluated using the scoring function SCORE (see Table 1). The best predicted affinity of the active form of PKB for ATP was nanomolar (pK = 9.5) while the pK was negative for the inactive form. This prediction was refined by the evaluation of all the 24 PKB–ATP complexes that kinDOCK can possibly build (from all known protein kinase–ATP complexes). The mean predicted affinity of those computed PKB–ATP complexes are micromolar (pK ∼ 7.1). On the contrary, the scores for the inactive form are all negative (mean pK ∼ −9.1), suggesting that ATP cannot fit into this conformation.
Table 1.
Discrimination of active/inactive forms by kinDOCK
| Best binder | ATP analog | |
|---|---|---|
| PKB active form (PDB1O6L) | 9.5 | 9.5 |
| PKB inactive form (PDB1GZN) | 3.5 | −3.2 |
The predicted affinity scores are given as −log(IC50) for the best ranked ligand as well as the best ATP analog.
In some protein kinases, the activation loop can adopt the so-called ‘DFG-out’ conformation and this structural rearrangement prevents the binding of most ligands, especially ATP and ATP-competitive inhibitors. This conformation can be stabilized by particular compounds binding deeply inside the protein core. The server kinDOCK can rapidly highlight such rearrangements and their implications in the ligand entrance. This is exemplified with the three distinct conformations of the protein kinase c-Kit. The latter has been co-crystallized with ATP, PP1 an ATP-competitive inhibitor (16) and STI-571 (also named imatinib mesylate or Gleevec) a ligand inducing a ‘DFG-out’ conformation. The corresponding structures have been submitted to kinDOCK and the affinity for the three different ligands was predicted (see results for human c-Kit in Table 2 and at http://abcis.cbs.cnrs.fr/LIGBASE_SERV_WEB/PHP/kindock_ex2.php). The predicted affinity of the active form of this protein (PDB1PKG) for ATP and PP1 is nanomolar while that for STI-571 is negative. The opposite situation is observed for the structure of c-Kit co-crystallized with STI-571 (PDB1T46) which adopts a ‘DFG-out’ conformation. A third structure of the c-Kit kinase which corresponds to an emtpy and inactive form does not bind to any of these ligands according to kinDOCK. This suggests that no unique structure can represent a protein kinase and that several conformations have to be determined or modelled.
Table 2.
Discrimination of distinct conformations of the c-Kit protein kinase by kinDOCK
| Best binder | ATP analog | STI-571 | PP1 | |
|---|---|---|---|---|
| c-Kit active form (PDB1PKG) | 10.6 | 8.7 | −6.8 | 8.0 |
| c-Kit inactive form (PDB1T45) | 4.9 | −9.0 | 1.1 | 2.4 |
| c-Kit bound to STI-571 (PDB1T46) | 9.3 | −3.8 | 8.9 | 9.1 |
The predicted affinity score of the best ATP analog, STI-571 and PP1 are shown. The affinity score deduced from experimental assays of c-Kit with STI-571 and PP1 are respectively ∼7.0 and ∼7.2 (16).
Poor scoring for a given type of ligand can also reveal incorrectly modelled side-chains or loops. This might allow the selection of optimal models prior to extensive VS in order to improve its hit rate (8). In order to evaluate the capabilities of kinDOCK to select the best model among a set of models for a given protein kinase, we use the example of the well-characterized CDK4 which has been, so far, recalcitrant to crystallization (17). A comparative docking was performed on the protein kinase CDK4 whose three-dimensional structure was modelled using various templates and the modelling server @TOME (http://abcis.cbs.cnrs.fr/atome/) in a fully automatic manner (see results at http://abcis.cbs.cnrs.fr/LIGBASE_SERV_WEB/PHP/kindock_ex.php). KinDOCK was used to discriminate three distinct models built from two closely related templates: CDK2 and CDK6. CDK6 and CDK2 are roughly 60 and 40% identical to CDK4. However, only CDK2 crystallized in an active form (in presence of ATP and cyclin A; PDB1QMZ) while both CDK2 and CDK6 can be observed in an inactive form (PDB1HCL and PDB1BLX, respectively). Models deduced from inactive forms failed to accommodate most ligands while the model built from the active CDK2 structure nicely accommodates ATP. To evaluate further the quality of these three models, we took advantage of the knowledge of two crystal structures of a mutated CDK2 with two known CDK4-specific inhibitors (17). The best model also provides us predicted affinities for these inhibitors (mean pK of ∼5.2, with the best model versus ∼1.3 and ∼1.6 with those from the two inactive forms) in agreement with the experimental ones (sub-micromolar range). The evaluation, by kinDOCK, of the crystal structures of the mutated CDK2 with these two inhibitors (PDB1GII and PDB1GIJ) provided similar values (predicted pK for their own ligand of ∼5.6 and 6.3, respectively). This result suggested that kinDOCK can correctly evaluate the active site conformation of molecular models while providing relatively accurate poses (see above) as well as relatively good estimates of protein–ligand interactions.
Focused screening
As illustrated above, kinDOCK allows a rapid screening of a small library of chemical compounds. Despite the partial loss of chemical diversity, the proposed approach focuses on privileged pharmacophores. The second advantage comes from the accuracy of the poses (see above) that allows for a better evaluation of protein–ligand interactions. This suggests that good starting hits can be detected using kinDOCK.
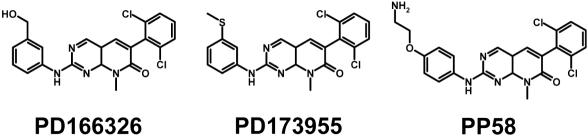
Alternatively, the specificity of a given inhibitor for a set of protein kinases can be evaluated by submitting each structure to kinDOCK. This might reveal potential side effects of the studied inhibitors (15,18). The question has also been recently addressed by experimental means for several protein kinase inhibitors (19). In the case of the pyrido[2,3-d]pyrimidine derivatives, affinity assays on the molecule PP58, closely related to the ligands PD166326 and PD173955 in our library (Figure 4), allowed us to evaluate the performance of kinDOCK. A good correlation between observed and predicted affinities was observed for a set of six distinct protein kinases (see Table 3). This result is in agreement with a specific amino-acid substitution (methionine versus threonine at the gatekeeper position) that distinguishes the two protein kinases subsets, with micromolar (pK score ∼6) versus nanomolar (pK score ∼9) affinities for the PP58 molecule. Similar correlations were observed with another set of protein kinases and two imidazol derivatives (data not shown). On the contrary, in the case of hymenialdisine, a correlation could not be found using kinDOCK, and no sequence variation in the active site was detected. Similar results have been described recently by others (15). Additional analyses will be necessary to explain the distinct binding affinities for this very rigid inhibitor. Similarly, some other parameters influencing binding energies cannot be evaluated directly by our approach (e.g. ligand flexibility, water displacement, metal binding,…).
Figure 4.
Structures of the compound PD166326, PD173955 and their analog PP58.
Table 3.
Discrimination of protein kinases with micromolar or nanomolar affinities for pyrido[2,3-d]pyrimidine derivatives by kinDOCK
| PD166326 (Predicted) | PD173955 (Predicted) | <PD> (Predicted) | pIC50(PP58) (Experimental) | |
|---|---|---|---|---|
| Mek1 bound to ATP (PDB1S9J) | 5.8 | 5.6 | 5.7 | 5.3 |
| Epha2r bound to AMP-PNP (PDB1MQB) | 5.7 | 6.7 | 6.2 | 5.4 |
| Ephb2r bound to adenine (PDB1JPA) | 5.7 | 8.5 | 7.1 | 8.0 |
| p38 inhibited form (PDB1DI9) | 5.0 | 10.0 | 7.5 | 8.3 |
| C-abl bound to STI-571 (PDB1IEP) | 8.5 | 9.8 | 9.1 | 9.3 |
| C-abl bound to PD173955 (PDB1M52) | 10.2 | 11.7 | 10.9 | 9.3 |
| Csk bound to staurosporine (PDB1BYG) | 10.2 | 10.5 | 10.3 | 9.4 |
The affinities of the PP58 (a pyrido[2,3-d]pyrimidine derivative related to PD166326 and PD173955) for each of the different protein kinases are deduced from experimental data (19). The mean predicted affinity score for the PD166326 and PD173955 ligands is shown in the <PD> column. The proteins in bold are the one with nanomolar affinities for PP58.
Conclusions and perspectives
Using kinDOCK, potential inhibitors can be rapidly identified for a given protein kinase. This approach may also rapidly reveal potential chemical substitutions to better fit the studied active site. This information can be used to filter the chemical libraries for virtual screening. The results presented herein, are promising, but more accurate affinity predictions could be obtained from the combined use of several evaluation programs.
As shown in the previous examples (described above), kinDOCK can discriminate active conformations from inactive ones, and the server works equally well for theoretical models and experimental structures. Using the server @TOME dedicated to molecular modelling, different conformations of a protein kinase can be automatically built and evaluated at a global and structural level. Then, the ATP-binding site of the various models can be evaluated at a local and functional level using kinDOCK. This approach might provide distinct conformations according to the template used (e.g. using related structures crystallized in ‘DFG-in’ or ‘DFG-out’ conformations). An alternative means for creating such variability in the active site structure was recently proposed and shown to improve VS results (20). In that work the diversity in the active site conformations is obtained by molecular dynamic simulations. In contrast, in the present work we use @TOME to model several conformations of the same active site in order to take into account the protein flexibility and a potential ligand-induced fit. We believe that our approach is much faster and will predict conformations more likely to be accessible, as they are already observed in related structures. The server kinDOCK will be connected with the modelling server @TOME in order to provide a complete and fully automatic pipeline for molecular modelling and focused screening based on protein structure similarity.
Acknowledgments
This study has been supported by the CNRS (‘ACI cibles thérapeutiques’). V.C. and L.M. are supported by the INSERM (Programme ‘Accueil Ingénieurs Grandes Ecoles’) and by the Ligue Contre Le Cancer, respectively. The authors wish to thank experimentalists for depositing crystal data to the PDB. The authors acknowledge the useful critics from Dr Cathy Royer. Funding to pay the Open Access publication charges for this article was provided by the CNRS.
Conflict of interest statement. None declared.
REFERENCES
- 1.Arora A., Scholar E.M. Role of tyrosine kinase inhibitors in cancer therapy. J. Pharmacol. Exp. Ther. 2005;315:971–979. doi: 10.1124/jpet.105.084145. [DOI] [PubMed] [Google Scholar]
- 2.Knighton D.R., Zheng J.H., Ten Eyck L.F., Ashford V.A., Xuong N.H., Taylor S.S., Sowadski J.M. Crystal structure of the catalytic subunit of cyclic adenosine monophosphate-dependent protein kinase. Science. 1991;253:407–414. doi: 10.1126/science.1862342. [DOI] [PubMed] [Google Scholar]
- 3.Baker D., Sali A. Protein structure prediction and structural genomics. Science. 2001;294:93–96. doi: 10.1126/science.1065659. [DOI] [PubMed] [Google Scholar]
- 4.Hare B.J., Walters W.P., Caron P.R., Bemis G.W. CORES: an automated method for generating three-dimensional models of protein/ligand complexes. J. Med. Chem. 2004;47:4731–4740. doi: 10.1021/jm0499054. [DOI] [PubMed] [Google Scholar]
- 5.Cohen-Gonsaud M., Catherinot V., Labesse G., Douguet D. From molecular modelling to drug design. In: Rychlewski K, editor. Progress in Nucleic Acid Research and Molecular Biology. Vol. 15. Springer-Verlag; 2004. pp. 35–71. [Google Scholar]
- 6.Shoichet B.K., McGovern S.L., Wei B., Irwin J.J. Lead discovery using molecular docking. Curr. Opin. Chem. Biol. 2002;6:439–446. doi: 10.1016/s1367-5931(02)00339-3. [DOI] [PubMed] [Google Scholar]
- 7.Evers A., Gohlke H., Klebe G. Ligand-supported homology modelling of protein binding-sites using knowledge-based potentials. J. Mol. Biol. 2003;334:327–345. doi: 10.1016/j.jmb.2003.09.032. [DOI] [PubMed] [Google Scholar]
- 8.Diller D.J., Li R.zz. Kinases, homology models, and high throughput docking. J. Med. Chem. 2003;46:4638–4647. doi: 10.1021/jm020503a. [DOI] [PubMed] [Google Scholar]
- 9.Noble M.E., Endicott J.A., Johnson L.N. Protein kinase inhibitors: insights into drug design from structure. Science. 2004;303:1800–1805. doi: 10.1126/science.1095920. [DOI] [PubMed] [Google Scholar]
- 10.Deshpande N., Addess K.J., Bluhm W.F., Merino-Ott J.C., Townsend-Merino W., Zhang Q., Knezevich C., Xie L., Chen L., Feng Z., et al. The RCSB Protein Data Bank: a redesigned query system and relational database based on the mmCIF schema. Nucleic Acids Res. 2005;33:D233–D237. doi: 10.1093/nar/gki057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hanks S.K., Quinn A.M. Protein kinase catalytic domain sequence database: identification of conserved features of primary structure and classification of family members. Meth. Enzymol. 1991;200:38–62. doi: 10.1016/0076-6879(91)00126-h. [DOI] [PubMed] [Google Scholar]
- 12.Catherinot V., Labesse G. ViTO: tool for refinement of protein sequence-structure alignments. Bioinformatics. 2004;20:3694–3696. doi: 10.1093/bioinformatics/bth429. [DOI] [PubMed] [Google Scholar]
- 13.Schuttelkopf A.W., van Aalten D.M. PRODRG: a tool for high-throughput crystallography of protein-ligand complexes. Acta. Crystallogr. D. Biol. Crystallogr. 2004;60:1355–1363. doi: 10.1107/S0907444904011679. [DOI] [PubMed] [Google Scholar]
- 14.Wang R., Liu L., Lai L., Tang Y. SCORE: a new empirical method for estimating the binding affinity of a protein–ligand complex. J. Mol. Model. 1998;4:379–394. [Google Scholar]
- 15.Rockey W.M., Elcock A.H. Rapid computational identification of the targets of protein kinase inhibitors. J. Med. Chem. 2005;48:4138–4152. doi: 10.1021/jm049461b. [DOI] [PubMed] [Google Scholar]
- 16.Tatton L., Morley G.M., Chopra R., Khwaja A. The Src-selective kinase inhibitor PP1 also inhibits Kit and Bcr-Abl tyrosine kinases. J. Biol. Chem. 2003;278:4847–4853. doi: 10.1074/jbc.M209321200. [DOI] [PubMed] [Google Scholar]
- 17.Ikuta M., Kamata K., Fukasawa K., Honma T., Machida T., Hirai H., Suzuki-Takahashi I., Hayama T., Nishimura S. Crystallographic approach to identification of cyclin-dependent kinase 4 (CDK4)-specific inhibitors by using CDK4 mimic CDK2 protein. J. Biol. Chem. 2001;276:27548–27554. doi: 10.1074/jbc.M102060200. [DOI] [PubMed] [Google Scholar]
- 18.Rockey W.M., Elcock A.H. Progress toward virtual screening for drug side effects. Proteins. 2002;48:664–671. doi: 10.1002/prot.10186. [DOI] [PubMed] [Google Scholar]
- 19.Wissing J., Godl K., Brehmer D., Blencke S., Weber M., Habenberger P., Stein-Gerlach M., Missio A., Cotten M., Muller S., et al. Chemical proteomic analysis reveals alternative modes of action for pyrido[2,3-d]pyrimidine kinase inhibitors. Mol. Cell Proteomics. 2004;3:1181–1193. doi: 10.1074/mcp.M400124-MCP200. [DOI] [PubMed] [Google Scholar]
- 20.Cavasotto C.N., Abagyan R.A. Protein flexibility in ligand docking and virtual screening to protein kinases. J. Mol. Biol. 2004;337:209–225. doi: 10.1016/j.jmb.2004.01.003. [DOI] [PubMed] [Google Scholar]