Abstract
The vast amount of protein sequence data now available, together with accumulating experimental knowledge of protein function, enables modeling of protein sequence and function evolution. The PANTHER database was designed to model evolutionary sequence–function relationships on a large scale. There are a number of applications for these data, and we have implemented web services that address three of them. The first is a protein classification service. Proteins can be classified, using only their amino acid sequences, to evolutionary groups at both the family and subfamily levels. Specific subfamilies, and often families, are further classified when possible according to their functions, including molecular function and the biological processes and pathways they participate in. The second application, then, is an expression data analysis service, where functional classification information can help find biological patterns in the data obtained from genome-wide experiments. The third application is a coding single-nucleotide polymorphism scoring service. In this case, information about evolutionarily related proteins is used to assess the likelihood of a deleterious effect on protein function arising from a single substitution at a specific amino acid position in the protein. All three web services are available at http://www.pantherdb.org/tools.
INTRODUCTION
The continued improvements in DNA sequencing technology are rapidly expanding our knowledge of the genomes and, by inference (through the genetic code and prediction of open reading frames), the proteomes of extant species. These DNA and protein sequences provide detailed information about molecular evolution. Combined with information about protein function derived from biochemical and genetic experiments, the molecular evolution data can shed light on the relationship between protein sequence and function. The PANTHER database (1,2) was designed to model the relationships between protein sequence and function for all major protein families, using molecular taxonomy tree building combined with human biological interpretation of the resulting trees. The trees are used to locate functional divergence events within protein families that define subfamilies of proteins of shared function.
The current version of PANTHER (6.0) contains trees for over 5000 protein families, divided into over 30 000 functional subfamilies. For each family and subfamily group, a multiple sequence alignment is constructed that aligns ‘equivalent’ positions (i.e. descended from the same ancestral codon) in each of the proteins in the group. Each multiple sequence alignment is then represented as a hidden Markov model (HMM) that summarizes, for each position, the probabilities of each of the 20 amino acids appearing (or of insertions and deletions) at that position in the given group of related sequences.
The resulting HMM parameters can be used in a number of scientific applications. We discuss two here. The first is classification of new sequences. The match between a sequence and an HMM is given a score by calculating the probability that the sequence was ‘generated’ by that HMM, and comparing it with the probability that the sequence was generated by a random HMM of the same length (3). For a new sequence, this HMM ‘score’ can be calculated for each of the family and subfamily HMMs, and the sequence is classified as belonging to same group as the best-scoring HMM (provided that the score is also statistically significant). In PANTHER, because each HMM is classified by the functions of its constituent proteins, protein sequences can be assigned to functional classes in a robust and consistent manner across different genomes. Currently, there are three different functional classifications associated with PANTHER HMMs: molecular function, biological process and component in a known metabolic or signaling pathway. The molecular function and biological process terms are largely a subset of those in the Gene Ontology (4), while the metabolic and signaling pathway components are taken primarily from the scientific literature (2).
The PANTHER HMMs, then, can be used to categorize genes (via their protein products) into functional categories robustly and comprehensively. Currently >75% of human proteins [represented by the ‘reviewed’ entries in the RefSeq database (5)] are classified into molecular function categories, a similar percentage to biological process categories, and ∼25% to components in canonical pathways. Functional groupings of genes have been used for statistical analysis of the results of genome-scale experiments, such as gene expression analysis (6,7), protein expression analysis (8) and even analysis of evolutionary selection pressure over large numbers of genes (9).
Another use of statistical models of protein families and subfamilies is in assessing the likely functional effects of non-synonymous single-nucleotide polymorphisms (nsSNPs) (1,10). In this case, patterns of sequence conservation and divergence found in related proteins from different organisms, or other genes in the same organisms, can be used to suggest amino acids that are important for function even for variants of the same gene in different individuals (polymorphisms). Amino acids that are conserved in related proteins despite ample time to diverge are likely to be critical for function. Simple conservation, where the same amino acid is found in all related proteins, is only the limiting case of non-random amino acid ‘profiles’ (11). Other profiles may arise from conservation of charge, hydrophobicity, or a number of other physico-chemical properties of amino acids, or from other functional or evolutionary constraints. These non-random profiles can be used to ‘score’ the likelihood of amino acid substitution to impact function, in a position-specific manner (i.e. different positions will have different amino acid profiles across the set of related sequences).
WEB SERVICES
There are three services currently available in the ‘Tools’ section of the PANTHER website (http://www.pantherdb.org/tools): the protein sequence classification service, the expression data analysis service and the nsSNP scoring service. Before describing these services in detail, for those already familiar with the PANTHER website we first list the newest features.
New features
Since the previous publication mentioning the interactive tools at the PANTHER website [Mi et al., (2)], several new features have been added to the PANTHER web services.
The expression data analysis service can now be used for any set of genes or proteins from any organism, not just the organisms stored on the PANTHER website (human, mouse, rat and Drosophila melanogaster). Users interested in analyzing other datasets, such as protein expression data mapped to UniProt, or gene expression data in canines or yeast, can now do so, using the new ‘PANTHER generic mapping file’ format described below. This format enables users to upload any proteins or genes, after scoring the associated protein sequences against the PANTHER HMM library using the scoring script available at http://www.pantherdb.org/downloads.
The expression data analysis statistics now include a Bonferroni correction for multiple testing. The Bonferroni correction is important because we are performing many statistical tests (one for each pathway or each ontology term) at the same time. This correction multiplies the single-test P-value by the number of independent tests to obtain an expected error rate. For pathways, we now correct the reported P-values by multiplying by the number of pathways tested. Some proteins participate in multiple pathways, so the tests are not completely independent of each other and the Bonferroni correction is conservative. For ontology terms, the simple Bonferroni correction becomes extremely conservative because parent (more general) and child (more specific) terms are not independent at all: any gene or protein associated with a child term is also associated with the parent (and grandparent, etc.) terms as well. We therefore modify the Bonferroni correction to account for the nesting of child terms below parent terms. Because at more specific levels it takes a larger number of independent terms to span the ontology, the number of independent tests can be seen as depending on the level of specificity in the ontology. Level 1 terms are treated as independent of other level 1 terms, but since all level 2 and 3 terms are subsumed by all of the level 1 terms they are not independent tests. We apply the same basic idea for lower level terms, though we need to adjust the count slightly to span the ontology, since not all level 1 terms are subdivided into level 2 terms, nor all level 2 terms into level 3 terms. For example, for level 2 terms, we treat each test as independent of the tests for other level 2 terms, and as independent of any level 1 terms that have no children.
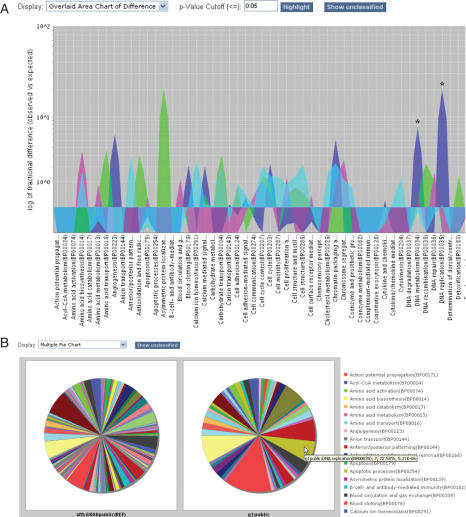
Users can now visualize the data underlying the binomial test statistics (see below). The counts of genes or proteins in each functional grouping can be viewed as pie or bar charts, and fold differences from the reference list can be graphed with statistically significant differences highlighted (Figure 1).
Figure 1.
Viewing data underlying statistical analysis of gene lists with respect to function. The data are from Cho et al. (16); each list comprises genes up-regulated at a given stage of the human cell cycle. (A) Overlay chart of four datasets (M, G1, S and G2 phases of cell cycle): logarithm of fold differences (compared with reference list) of the numbers of genes in each biological process category. Each dataset is shown in a different color (G1 blue, S magenta, G2 green, M light blue); statistically significant differences are indicated with an asterisk. (B) Multiple pie chart of the biological processes represented in the reference list (left) versus the list of genes up-regulated during G1 phase (right). Mousing over a pie slice shows details about the comparison with the reference list; in this example, among the genes up-regulated in G1 (right) there are many more genes involved in DNA replication than expected by chance (the same color slice in the reference list at left).
The coding SNP scoring service now provides a link to a combined view of the family tree and the multiple sequence alignment, where the data used to calculate the amino acid probabilities are highlighted. The column of the multiple alignment that corresponds to the position of the amino acid substitution is highlighted in red, while the subtree over which the amino acid is conserved is highlighted in gray (Figure 2). In addition, the subPSEC score (1) is now converted to a more readily interpretable probability of deleterious effect on protein function, Pdeleterious, as described below.
Figure 2.
Graphical view of the evolutionary data used to calculate coding SNP scores. The multiple sequence alignment of UniProt sequences (right) is displayed next to the protein family tree that shows the relationships between functionally distinct subfamilies. In this example, the uploaded sequence was for the product of the ABCA1 gene (RefSeq NP_005493), for the mutation L1075V. The column corresponding to the substituted amino acid is highlighted in red, and the subfamilies (ABCA1, ABCA4, ABCA7) used to calculate the score Pdeleterious are expanded in the tree view on the left. See text for more details. The user can expand and collapse tree nodes by clicking on any node (green circles or blue diamonds indicating subfamily nodes). Other subfamilies (e.g. ABCA2, ABCA12, ABCA13) are shown collapsed here.
Finally, a new version of the PANTHER library (6.0) has been released. All protein subfamilies have been reviewed and updated by expert curators, who have also updated and corrected the ontology terms. UniProt sequences are now used for building the library of families, simplifying the links to detailed functional information for individual proteins. The HMMs underlying the sequence classification service and coding SNP scoring statistics therefore incorporate the most recent protein sequence and annotation data. Version 1.2 of PANTHER Pathway has also been released, containing 107 pathways (primarily signaling pathways) that can be viewed interactively.
Protein sequence classification service
The protein sequence classification service is available at http://www.pantherdb.org/tools/hmmScoreForm.jsp. The user inputs a protein sequence (as a string of amino acid single letter codes or FASTA format) and presses the ‘submit’ button. The sequence is then scored against the entire PANTHER ‘library’ of family and subfamily HMMs (1), and if the top hit is statistically significant (E-value < 0.001), that hit is returned. The alignment and E-value (Bonferroni-corrected P-value for the match) is shown, as well as the HMM name and links to additional information about the HMM, including the molecular function, biological process and pathway component associations, and the training sequences used to build the HMM and the protein sequence family tree.
The top scoring HMM can be either a family or subfamily HMM. Family HMMs are generally associated with less specific functional information than subfamilies. The E-value of the hit is also important for interpreting the results, and in addition to the actual E-value, we provide a simple icon that shows the empirically derived confidence level of the classification. The icon shows three filled circles if the classification confidence level is high (E-value < 10−23), two filled circles if the confidence is medium (E-value < 10−11), and one filled circle if the confidence level is low. More detailed information is available by clicking on the help icon on the results page.
The PANTHER website also allows access to pre-calculated HMM scoring results for the complete proteomes derived from the human, mouse, rat and D.melanogaster genomes. These data can be accessed using the text search box on the home page, or the batch search page accessible from the Genes section of the website (http://www.pantherdb.org/genes).
Tools for finding statistically significant functional associations in genomic experimental data (Expression data analysis service)
There are two tools available in this section of the website (http://www.pantherdb.org/tools/genexAnalysis.jsp). Both are designed to uncover statistically significant relationships between input data and gene or protein functions. The main applications for this type of analysis has been for finding functional trends in mRNA microarray data or protein expression data from mass spectrometry, although it has been used to aid in data interpretation from a number of genome-wide studies such as gene essentiality screens (12) and comparative genomics studies such as tests for positive selection across many genes (9).
The first tool is for analysis of gene or protein lists with respect to function. The test is the conceptually simple binomial test described in reference (6). Each input list is divided into groups based on the functional classification (either molecular function, biological process, or pathway). A reference list (all of the genes/proteins from which the list was drawn) is divided into groups in the same way. Then, for each functional category, the binomial test is applied to determine whether there is a statistical over- or under-representation of genes/proteins in the input list relative to the reference list.
The input is from one to four lists of genes or proteins (plus, optionally, a ‘reference list’), which are uploaded onto the website. These lists are not stored on the site once the user ends the session. One of two formats must be used, depending on whether the user wishes to use the pre-calculated HMM scoring data stored on the PANTHER website, or to use a file they have generated by scoring their own protein sequence set against the PANTHER HMMs (available for download at http://www.pantherdb.org/downloads). For using the pre-calculated PANTHER classification data, the format is simply a single column file of identifiers that can specify records in the PANTHER database. Currently the pre-calculated data covers only the human, mouse, rat and Drosophila genomes. The supported identifiers are listed on the list upload page, but they include gene identifiers [Entrez Gene (5) identifiers for human, mouse and rat, or FlyBase (13) FBgn numbers for Drosophila], protein identifiers (RefSeq or FlyBase) and gene symbols. For the user-generated data, the ‘PANTHER generic mapping file’ format must be used instead. This format consists of two columns: the first column is an arbitrary identifier that the website will temporarily store (again only for the session) which allows the user to uniquely specify each record in the dataset, so they can track that identifier on the website; the second column is the PANTHER HMM identifier (e.g. PTHR19266, or PTHR19266:SF40), which is used to look up molecular function, biological process and pathway associations.
The output of the tool is a list of P-values for under- or over-representation of each functional category in each of the input lists. From this output page, the user can export the statistics, or follow links to graphically view (as pie charts or bar graphs) the data that were used to compute the P-values, or to look at the list of genes/proteins in any functional group. When pathways are chosen as the functional categories, clicking on the pathway name brings up pathway diagrams colored according to preferences specified by the user.
The second tool in this section is for analysis of a complete list of genes/proteins that have numerical data associated with each gene/protein. The most commonly used numerical data are probably the fold-change value for each gene in a differential expression experiment, but the statistical test is general enough to handle any numerical data, continuous or discontinuous. The statistical tool builds a distribution of values for all input data in the list (this becomes the reference distribution), and then divides the input data into functional categories and builds a distribution of values for each category. The probability that the functional category distribution was drawn randomly from the reference distribution is estimated using the Mann–Whitney Rank-Sum Test (U-test), as described in (9). Using the whole distribution of values has been shown (9,14) to provide a more sensitive test than the simple list-based test described above.
For the numerical data test, the user inputs a single file. Like the list comparison tool, there are two formats for the uploaded file, depending on the desired source of the PANTHER classification data: either the pre-calculated classifications available on the PANTHER site, or a user-generated file. For using the pre-calculated PANTHER data, the file must contain two columns: the first is the gene or protein identifier, and the second is the numerical value. For user-specified data, the file must contain three columns: an arbitrary tracking identifier (e.g. a UniProt identifier or gene symbol); the PANTHER HMM identifier indicating the classification of the gene/protein; and the numerical value.
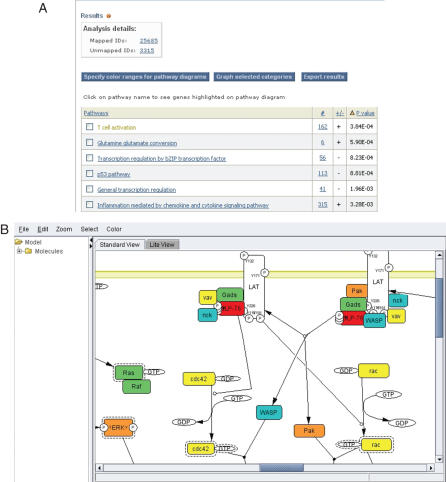
The output of the tool is a list of P-values for each comparison between a functional category distribution and the reference distribution. Each distribution, and how it compares with the reference distribution, can be viewed graphically from the output page. We find that this is critical for interpreting the any deviation between the functional category distribution and the overall distribution. The genes/proteins in each category can also be viewed from the output page by clicking on the listed counts. In addition, for pathways, clicking on the pathway name will bring up an interactive Java applet that colors the pathway using a ‘heat map’ derived from the input values (Figure 3).
Figure 3.
Expression data analysis and visualization on the PANTHER website. (A) Mann–Whitney U-test results, and (B) CellDesigner (15) diagram of the T-cell activation signaling pathway from the PANTHER Pathway database (accession P00053, author Adam Douglass). This applet colors proteins according to a ‘heat map’ calculated from user-input values. Protein components are mapped to PANTHER HMMs. Active forms (dashed-line boxes) and phosphorylated forms (small circles around the letter ‘P’) of proteins are clearly indicated in the diagram. A total of 107 pathways (mostly signaling pathways) are currently available.
Coding SNP scoring service
The non-synonymous SNP scoring service is available at http://www.pantherdb.org/tools/csnpScoreForm.jsp. The methodology used to generate the scores is described in detail in (1) and summarized in (14). Briefly, the method uses a multiple alignment of a family of protein sequences, together with information about functional subfamilies within that family, to estimate the probabilities of different amino acids occurring at different positions in the protein family. High probability amino acids are likely to result in a functional protein, while low probability amino acids are likely to have a deleterious effect on protein function. We quantify the likely functional effect with a substitution position-specific evolutionary conservation (subPSEC) score, calculated as simply the log of the ratio of the probabilities of the two substituted amino acids: ln(Psub/Pwt), where Psub is the probability of the substituted amino acid and Pwt is the probability of the wild-type amino acid. Smaller (more negative) subPSEC scores indicate a higher likelihood of being deleterious. We have recently added a third parameter to the subPSEC score: the number of independent counts nic, a measure of the (global) diversity of sequences over which a position has been conserved. In effect, this parameter gives a greater probability of functional impact for positions that have been conserved over greater evolutionary distances. Based on calibration using a large set of known disease-causing non-synonymous mutations, as well a large set of randomly sampled non-synonymous human SNPs (1), we can express the probability of a nsSNP being deleterious (Pdeleterious) as a function of the subPSEC score [details are given in reference (17)]:
in which subPSEC = −0.88lnPa + 0.89lnPb − 0.94lnnic, where Pa is the larger and Pb the smaller, of the two amino acid probabilities, and nic is the number of independent counts. Pa, Pb and nic are all calculated in a position-specific manner, using the largest subtree of the family tree that both (1) conserves the same amino acid as the input sequence and (2) contains the PANTHER subfamily that had the best score to the input sequence.
To use the coding SNP scoring service, there are two boxes on the input form that must be filled out. The first is the sequence of the protein, in the same one-letter code format as for the classification service above. The second is a list of amino acid substitutions, in the standard mutation notation, e.g. D432A, where the wild-type amino acid is D (D must appear at position 432 in the sequence entered into the first box), and A is the substituted amino acid. Multiple substitutions may be entered into the second box, separated by a <return> character. The exact number of substitutions that can be handled per query (i.e. before the page times out) ranges from a minimum of 10 to a maximum of hundreds, depending on the length of the query protein sequence and the size of the PANTHER family it matches.
The wild-type protein sequence is then searched against the PANTHER HMM library to find the highest-scoring (statistically significant) PANTHER HMM, using the same methods as the classification service above. This search specifies the multiple sequence alignment, subfamily (if possible) and tree that will be used to estimate the substitution probabilities, and also specifies the position of the substitution in the multiple alignment (by aligning the user's sequence to the existing multiple alignment).
The output is a list of the substitutions entered by the user, with the amino acid probabilities derived from the multiple sequence alignment, Pwt and Psub, nic, the subPSEC score, and the predicted probability that the substitution is deleterious, Pdeleterious. The alignment and tree used to generate the amino acid probabilities can be viewed by clicking on the link from the ‘position’ column of the output data. If the substitution occurs at a position that does not appear in the multiple alignment, a subPSEC score cannot be generated and the output will return the text string ‘does not align to HMM’, indicating that the substitution occurs at a position that is inserted relative to the consensus HMM for the given family. In most cases, these positions are not modeled by the HMMs simply because they do not appear in most of the related sequences; as a result, substitutions at inserted positions are not generally likely to be deleterious.
Acknowledgments
The work was supported by Applied Biosystems, and funding for the Open Access publication was provided by SRI International.
Conflict of interest statement. None declared.
REFERENCES
- 1.Thomas P.D., Campbell M.C., Kejariwal A., Mi H., Karlak B., Daverman R., Diemer K., Muruganujan A., Narechania A. PANTHER: a library of protein families and subfamilies indexed by function. Genome Res. 2003;13:2129–2141. doi: 10.1101/gr.772403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mi H., Lazareva-Ulitsky B., Loo R., Kejariwal A., Vandergriff J., Rabkin S., Guo N., Muruganujan A., Doremieux O., Campbell M.J., et al. The PANTHER database of protein families, subfamilies, functions and pathways. Nucleic Acids Res. 2005;33:D284–D288. doi: 10.1093/nar/gki078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Eddy S.R. Hidden Markov models. Curr. Opin. Struct. Biol. 1996;6:361–365. doi: 10.1016/s0959-440x(96)80056-x. [DOI] [PubMed] [Google Scholar]
- 4.Gene Ontology Consortium. Gene ontology: tool for the unification of biology. Nat. Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pruitt K.D., Maglott D.R. RefSeq and LocusLink: NCBI gene-centered resources. Nucleic Acids Res. 2001;29:137–140. doi: 10.1093/nar/29.1.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cho R.J., Campbell M.J. Transcription, genomes, function. Trends Genet. 2000;16:409–415. doi: 10.1016/s0168-9525(00)02065-5. [DOI] [PubMed] [Google Scholar]
- 7.Maere S., Heymans K., Kuiper M. BiNGO: a Cytoscape plugin to assess overrepresentation of gene ontology categories in biological networks. Bioinformatics. 2005;21:3448–3449. doi: 10.1093/bioinformatics/bti551. [DOI] [PubMed] [Google Scholar]
- 8.Freeman W.M., Brebner K., Amara S.G., Reed M.S., Pohl J., Phillips A.G. Distinct proteomic profiles of amphetamine self-administration transitional states. Pharmacogenomics J. 2005;5:203–214. doi: 10.1038/sj.tpj.6500309. [DOI] [PubMed] [Google Scholar]
- 9.Clark A.G., Glanowski S., Nielsen R., Thomas P.D., Kejariwal A., Todd M.J., Tanenbaum D.M., Civello D., Lu F., Murphy B., et al. Inferring nonneutral evolution from human–chimp–mouse orthologous trios. Science. 2003;302:1960–1963. doi: 10.1126/science.1088821. [DOI] [PubMed] [Google Scholar]
- 10.Thomas P.D., Kejariwal A. Coding single nucleotide polymorphisms associated with complex versus Mendelian disease: evolutionary evidence for differences in molecular effects. Proc. Natl Acad. Sci. USA. 2004;101:15398–15403. doi: 10.1073/pnas.0404380101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gribskov M., McLachlan A.D., Eisenberg D. Profile analysis: detection of distantly related proteins. Proc. Natl Acad. Sci. USA. 1987;84:4355–4358. doi: 10.1073/pnas.84.13.4355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yu H., Greenbaum D., Xin Lu H., Zhu X., Gerstein M. Genomic analysis of essentiality within protein networks. Trends Genet. 2004;20:227–31. doi: 10.1016/j.tig.2004.04.008. [DOI] [PubMed] [Google Scholar]
- 13.FlyBase Consortium. The FlyBase database of the Drosophila genome projects and community literature. Nucleic Acids Res. 2002;30:106–108. doi: 10.1093/nar/30.1.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mootha V.K., Lepage P., Miller K., Bunkenborg J., Reich M., Hjerrild M., Delmonte T., Villeneuve A., Sladek R., Xu F., et al. Identification of a gene causing human cytochrome c oxidase deficiency by integrative genomics. Proc. Natl Acad. Sci. USA. 2003;100:605–610. doi: 10.1073/pnas.242716699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Funahashi A., Morohashi M., Kitano H. CellDesigner: a process diagram editor for gene-regulatory and biochemical networks. Biosilico. 2003;1:159–162. [Google Scholar]
- 16.Cho R.J., Huang M., Campbell M.J., Dong H., Steinmetz L., Sapinoso L., Hampton G., Elledge S.J., Davis R.W., Lockhart D.J. Transcriptional regulation and function during the human cell cycle. Nature Genet. 2001;27:48–54. doi: 10.1038/83751. [DOI] [PubMed] [Google Scholar]
- 17.Brunham L.R., Singaraja R.R., Pape T.D., Kejariwal A., Thomas P.D., Hayden M.R. Accurate prediction of the functional significance of single nucleotide polymorphisms and mutations in the ABCA1 gene. PLoS Genet. 2005;1:e83. doi: 10.1371/journal.pgen.0010083. [DOI] [PMC free article] [PubMed] [Google Scholar]