Abstract
The massive production of biological data by means of highly parallel devices like microarrays for gene expression has paved the way to new possible approaches in molecular genetics. Among them the possibility of inferring biological answers by querying large amounts of expression data. Based on this principle, we present here TOM, a web-based resource for the efficient extraction of candidate genes for hereditary diseases. The service requires the previous knowledge of at least another gene responsible for the disease and the linkage area, or else of two disease associated genetic intervals. The algorithm uses the information stored in public resources, including mapping, expression and functional databases. Given the queries, TOM will select and list one or more candidate genes. This approach allows the geneticist to bypass the costly and time consuming tracing of genetic markers through entire families and might improve the chance of identifying disease genes, particularly for rare diseases. We present here the tool and the results obtained on known benchmark and on hereditary predisposition to familial thyroid cancer. Our algorithm is available at http://www-micrel.deis.unibo.it/~tom/.
INTRODUCTION
Ceaseless advances in biotechnology, along with the growing experience cumulated by researchers in recent years, has allowed a continuous and faster blooming of the number of genomes being sequenced and, most importantly, annotated. The consequent necessities of storing, retrieving, sharing and, in particular, understanding this vast amount of data led to the creation of genome databases, an open source of genetic information for scientists worldwide.
Whereas the genomic era opened the doors to the very existence of such large and comprehensive (omic) data repositories, the strongest urgency of the post-genomic era is now to interrelate various sources of biomedical information.
Several parallel efforts are currently underway to achieve a better understanding of the human genome. These actions are turned to the extraction of high-throughput information from global approaches such as the International HapMap Project (1) for identification of single nucleotide polymorphisms or the prosecution of the ENCODE [ENcyclopedia Of DNA Elements (2)] for the identification of all functional elements in the genome sequence. Therefore the integration of various existing and upcoming efforts is going to be a key element for the full comprehension of the cellular machinery.
We focus here on one of the many fields that will strongly benefit from such an integration: the study of hereditary diseases. Often more than one gene is involved in life threatening misfunctioning of cellular functions. To characterize such diseases the identification of all the responsible genes is eventually a crucial requirement. This process usually involves costly, time consuming and difficult tracing of large family lineages to follow the line of transmission of genes and thus to define the linkage areas where genes responsible for the disease could be located.
Computational technologies can appropriately be employed to integrate available data and can, in principle, be used to save on the expensive process of candidate genes selection. In this article we describe TOM [Transcriptomics of OMIM, (3)], an automated pipeline for the extraction of the best candidate genes for a given genetic disease. The procedure is based on two possible starting points. On one instance the accepted input is a list of one or more genes (called the seed(s) of our search) plus the chromosomal area where the unknown gene is located. This option (One Locus option) is suited for cases where the disease is minimally characterized and at least one responsible gene is known. The algorithm performs the necessary steps to extract from the linkage area, listed in the input, the genes that have the highest chances of being functionally related to the seeds. The second option (Two Loci option) is designed for poorly characterized diseases, when no specific gene is a priori known. At least two bona fide linkage areas need to be present. It is therefore possible to query the two genome tracts associated to the same pathology. The algorithm extracts the lists of genes annotated on each linkage region and searches for pairs that have similar expression or functional profiles.
The scientific rationale behind TOM is rooted on three characteristic gene features: gene mapping, expression profiling and functional annotations. The combination of these three features enables the selection of genes that have desirable characteristics, and meanwhile the filtering of possible candidates that do not share them.
The first step, gene mapping, often a bottleneck in past times, is now inherent to the human DNA sequence. Since the decoding of the human genome, it is in fact possible to select genes matching any specific area of the genome. The definition of one or two genome regions of interest represents obviously the first selection criterion for the candidates to a given disease. TOM then proceeds to retrieve the list of annotated genes that are located in the regions.
Second, genes that have a common transcriptional regulation (resulting in coherent expression profiles), also have a high likelihood of being involved in the same cellular process (4–7). Expression profiling allows to record the activation/silencing of number of genes across different experiments or conditions and it is most commonly performed by means of DNA microarrays. Measuring with appropriate metrics the distance in the transcriptional profiles (i.e. the mRNA levels of two or more genes across a set of conditions) enables the identification of gene sets sharing similar behaviors. Applying this second selection criterion to the queried genetic interval(s), and extracting only the messengers with significant correlation to the seeds, leads to a strong reduction of the list of candidates.
Finally, proteins involved in the seed's cellular processes are likely to be encoded by candidate genes as well. Functional analysis consists in characterizing a gene's cellular role. Functional views of a protein can be obtained using the controlled vocabulary defined by the Gene Ontology Consortium [GO (8)]. The GO in fact enables the identification of a gene's molecular function and, in particular, of the biological process/processes in which it is involved. This third and final selection step then focuses one more time on the potentially relevant genes, selecting only those that are functionally related to the seeds (One Locus) or that share the same function/s (Two Loci). To allow more flexibility to the user, it is possible to disjoin the second and third step.
Dealing with the human genome represents a complex challenge and several efforts have been undertaken with the specific aim of facilitating the understanding of genetic diseases, namely the extraction of candidate gene lists for a given disease. These approaches can be divided in two broad categories, one mainly based on the use of ontologies (structured vocabularies for the classification of items) and the other that relies more on structural characteristics of the genes or their products. For both approaches several interesting strategies have been implemented. In the first category, approaches range from (i) the estimate of the association between terms from controlled vocabularies, that define a disease, and genomic sequences associated in literature to (ii) the evaluation of similarity among genes based on phenotypic descriptions (9,10). In the second category algorithms (11,12) are based on structural characteristics such as protein size or degree of conservation across evolution. Other approaches rely on the information obtained performing composite queries across several databases, including mouse and human gene expression (13). Besides their meaningful rationale, both approaches also have their disadvantages. The first class suffers from incompleteness owing to the still ongoing annotation process. The second class makes use of characteristics that are less biased than annotations, and relies on rules that have broad applicability, but may miss the specificity of the gene-by-gene discovery that is conversely available through ontologies.
TOM de facto merges the strategies used by the different approaches described above and its main advantage can be defined in its flexibility. On one hand, it is not strictly designed for queries on disease-related genes, but can be used for any type of gene(s)–locus or locus–locus inquiry. Furthermore, it can be used both by an investigator with a good level of a priori knowledge on the disease (One Locus option) and when the research is still at early stages (Two Loci option). Finally, the user can modulate the filtering ability, since he can decide whether to apply unsupervised expression neighborhood analysis, taking advantage of the unbiased transcriptional information, or the functional annotation, relying on the careful curation of the GO vocabulary.
We describe here in details the differences between TOM and two other applications that, among the published works, appear to be more related to our approach. The work of Tiffin et al. (14) makes use of gene expression information and constitutes an interesting approach based on the association (performed on Medline abstracts text-mining) between diseases and anatomy terms, based on frequency of occurrence. The top ranking associations are then used to mine ENSEMBL (http://www.ensembl.org) in search of the disease genes annotated to the corresponding anatomic sites. This approach lacks three features that are advantageously implemented in TOM: first it is presented as a method and not offered as an online service to geneticists; second, gene expression data are obtained as ENSEMBL annotation, thus generating the drawbacks described above for the annotation methods and third, TOM extracts genes coexpressed in any anatomic site or tissue, in diseased and non diseased samples. This opportunity allows a broader search and the identification of genes potentially difficult to capture in the disregulated processes, but that still share strong synergistic activities, present in the normal status.
A second program named POCUS, designed by Turner et al. (15) takes advantage of a sophisticated use of both GO and InterPro domain (16), evaluating the enrichment for any annotation of a set of genes localized on two given chromosomal areas defined by the user. The enrichment represents an indicator of commonalities among the genes. This approach is partially related to our Two Loci option, offering the advantage of allowing researches with very little a priori knowledge. However, it misses two issues addressed by TOM: (i) our One Locus option allows a more focused and targeted approach for better known diseases and (ii) Turner's approach may miss information that could be obtained by mining on other databases. In fact the authors state that the quality and number of annotation is biased by the number and quality of previous findings made on the genes (that appear as annotations IDs).
Finally, a very recent work from Franke et al. (17) approaches the problem of the relationships among genes of interest that can in some cases overlap with hereditary disease research. This work describes a complete integrated approach, making use of Bayesian networks, based on GO, gene expression and protein–protein interaction, as well as pathways and information from human yeast two hybrid (Y2H) experiments (18). This tool allows the prediction of genes that are functionally related to candidate chromosomal areas and is thus related to the Two Loci approach described in TOM. The interesting strategy described in this paper could then be used to integrate our tool.
In conclusion, we believe that TOM, which parallels the successful work of Mootha et al. (19) who devised an integrative approach to gain insight into cytochrome c oxidase deficiency, can be a valuable and efficient resource to help in genes extraction. Certainly, more options can be integrated to take better advantages of the existing but partial information sources, i.e. to detect unpredicted or non-coding genes.
MATERIALS AND METHODS
The Seeds of the search
The input data to TOM can be constituted by a (list of) gene(s) and one or two chromosomal areas of interest. In the One Locus option, while the chromosomal area represents the hypothesis to test, the input gene(s) are the queries of the search. For this reason almost invariably, but not exclusively, the seed of the search will belong to the repository of the Online Mendelian Inheritance in Man (OMIM) database. This repository stores a comprehensive collection of genes known to be related to human diseases. OMIM is updated daily and stores information (mainly genes) related to disorders inherited in a Mendelian manner, where traits are passed from parents to children. Any type of gene can be used with this method. Nevertheless, because most of the power in our procedure is given by expression data, we named the application TOM (Transcriptome of OMIM).
The three-step filtering algorithm
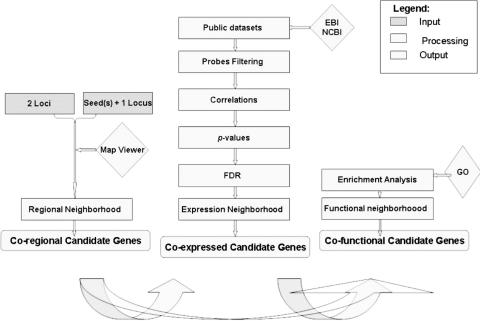
We describe here the detailed steps allowing the selection of the final candidate gene sets for an hereditary disease (see Figure 1).
Figure 1.
Global description of the process. The three steps of the algorithm, along with the databases and the intermediate and final results are shown in the figure. The output can be used at the end of the second step, in the form of co-expressed genes, or refined through the third step where the functional analysis (based on GO) is performed. The longest arrow depicts the alternate route to the functional analysis.
The first step is designed to select the list of genes mapped on the chromosomal area/s of interest, using genome sequence information.
Then, in the second step, TOM employs transcriptome data from public repositories. TOM retains here only the genes that have related expression variations in the datasets, either among them (Two Loci) or to the seeds (One Locus). Formally, this is achieved defining the expression neighborhood, i.e. the set of genes encoded in the genomic area of interest that are related among them or to the seeds, based on the similarity of their expression. We define a metric space M = (X,ρ), where X are vectors of expression values a gene assumes on a given sorted set of experiments (activation profile) and ρ the Spearman rank coefficient of correlation (20), a non-parametric measure of data trend correlation based on rankings. We then define the set N as the expression neighborhood of a gene g if there exists an open ball with center s and radius r, , where the radius r was defined by means of statistical significance. Namely, we evaluate the P-values of the correlation tests and select the genes whose correlation value is significant at a given value of rejection. Evaluation of P-values is performed assuming that the correlation values are distributed using Student's t cumulative distribution, with a number of degrees of freedom corresponding to the number of samples in the microarray experiment.
Given the high number of correlation tests performed in TOM, P-values are corrected for multiple testing by using the false detection rate (FDR), as defined by Ref. (21). FDR controls a different probability than that which is controlled with the better known P-value. In fact, P-values control the number of false positive over the number of truly null tests, while FDR controls the number of false positive over the number of significant tests. Several ways of estimating this number have been proposed, we adopted the solution devised by Tom Nichols (see http://froi.sourceforge.net/documents/technical/matlab/FDR.html), that rescales the P-value obtained on a single test multiplying it by a combination of indexes related to the total number of tests performed: , where pi represents the i-th of the total K single P-values. Correction was performed on a seed by seed basis, this means that the genes in the seeds list or in the first chromosomal area are considered independent tests.
Finally, in the third optional step, we further filter candidates, based on their functional role(s). To this end we use GO [the more widespread controlled and hierarchically structured vocabulary for the description of genes and genes' products characteristics in any organism (8)] to better interpret the genes selected in the expression neighborhood, and extract genes related to the same biological process(es). In particular, we use the hypergeometric distribution
to evaluate the probability that in a sample of size n, r items of a given type—a type characterizing n1 items in a population of N items—can be selected without replacement (20). This probability statistically validates the proportion of genes in the group of candidate genes (enriched for some known function), compared with what would be expected by chance alone. The third step can alternatively be performed directly after the first step, simply by skipping the transcriptional analysis.
This statistically validated triple filtering allows the targeted extraction of a shortlist of candidate genes, thus saving resources for the following costly and time-consuming genetic analysis.
Implementation
The tool core is developed under R and the user interface is developed using Php. Users requests are initially stored in a database (MySql), where a batch scheduled task retrieves and processes them, while the user interface is waiting. For defining the position of the bands, we use the NCBI MapViewer (http://www.ncbi.nlm.nih.gov/mapview/). The BUILD.35.1 genome data are stored in the TOM database. Once the results are obtained, the website will access and display them on the user's browser. The two alternative types of input are accessed from two different web-pages. One Locus allows the algorithm to accept as input one or more gene symbols or Affymetrix id (separated by semicolons) plus a linkage region; Two Loci accepts two distinct linkage regions. Both pages allow the user to select the maximum P-value (corrected for multiple hypotheses) accepted for significance. Regions are input as chromosome numbers, arms and bands. All stored genes found in the region(s) are retrieved from the database and associated to the annotations provided by Affymetrix. These features are contained in the Annotation files provided by the NetAffx analysis center (http://www.affymetrix.com).
The stored microarrays can then be searched to retrieve the expression values for the correlation analysis by checking the Expression Correlation filtering check box and setting the FDR threshold field. Microarray expression values undergo a double filtering process on the basis of the calls (flexible filtering) and of a fold-change and absolute filter variation over samples (max/min < 3 and max − min < 100, fixed filtering). This double filtering is meant to allow a stringent and constant selection based on the variation of the expression profile, while preserving the maximum amount of information, based on the general quality of the array. The quality of the array is related to the number of present calls available. For this reason, we filtered the array spots based on the assumption that a minimum amount of information can be extracted from the arrays. Thus, while we performed a strong filtration for 95% present calls in better quality arrays, we accepted also 50% present calls in worse ones. This information is stored with the array name. This procedure leads to expression matrices whose final size is around ≤5000 probe sets. Additional quality control information is also stored for each candidate gene as the percentage of samples in which both probe set evaluated in the correlation were called present.
Experiments conducted on Affymetrix chips (Human Genome U133A, U95A, U133A and B and U1333plus2 chips with detection calls available) were downloaded from the repository of Gene Expression Ominbus (GEO, http://www.ncbi.nlm.nih.gov/geo/) and EBI repository ArrayExpress (http://www.ebi.ac.uk/arrayexpress/) using a Perl script to capture the expression values. We performed these results loading ∼40% of the human microarray experiments with detection calls available. The process of populating TOM is ongoing and will be updated on a monthly basis. Besides the expression values, samples description, detection, platforms, gene chip array, sample conditions and authors are also stored. These data are accessible in the Results page.
The functional analysis based on GO identifies distribution and can be performed by checking the Validate GO IDs check box. We applied the Biobase and GOstats bioconductor packages (22) to perform the functional analysis. This package computes the hypergeometric probability using as items the LocusLink identifiers (http://www.ncbi.nih.gov/entrez) of each Affymetrix probe of query results. Each LocusLink identifier belongs to several GO categories and the package counts the number of appearances of each GO term for the genes in the candidate group as well as in the whole pool of genes extracted from the chip (reference group). We modified the GOstats package so that it was possible to evaluate the P-value for a group composed by the union of all the sets of GO identifiers belonging to the Affymetrix chips involved in the user request.
Since running these tasks can require several minutes, an e-mail service is provided that sends to the user a request identifier as soon as the user request is stored. Results are then mailed in tabular format with the URL to their online version when the process is terminated. The assigned code also allows the user to retrieve results at later times, since they are stored for two weeks. It is also possible to assign a friendly user-defined name to the request for tasks management facilitation.
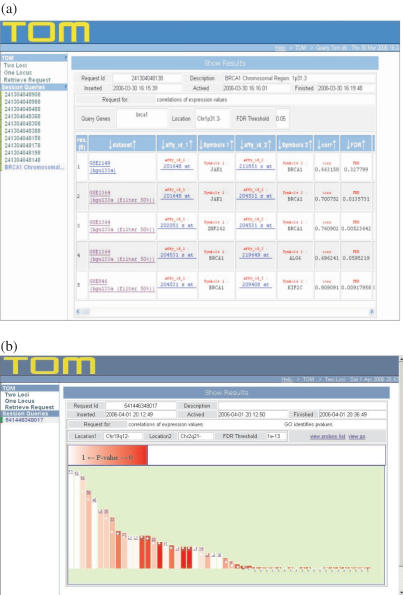
The output contains a matrix with information for each candidate gene and the corresponding seed (in case of One Locus option it is stored indifferently in the first or second column) related to the analyses performed such as correlation values, corresponding FDR, percentage of overlapping present samples for correlation evaluation, GOIDs, enrichment P-values for the three GO categories: Molecular Function, Biological Process and Cellular Component as well as several records from Affymetrix descriptive annotations (Figure 2a). The website finally offers an histogram showing a visual representation of the GO categories involved and their P-values (Figure 2b).
Figure 2.
(a) It shows an example of Results page for familial Breast Cancer, highlighting the known relationship between BRCA1 and JAK1. The Results table can scroll and more information, such as correlation values, then become visible. (b) It shows the graphical output of the functional analysis of Two Loci problem, namely Thyroid Cancer. This allows a rapid overview of the results. Every GO category of the candidate genes is represented in the bar graph. Categories are sorted based on the number of hits (height of the bar) and the P-value information is carried by a white to red shade of color. Tall white bars represent the best candidates, small red ones the worse.
RESULTS AND DISCUSSION
We present here some examples of the use of TOM for One Locus and Two Loci option.
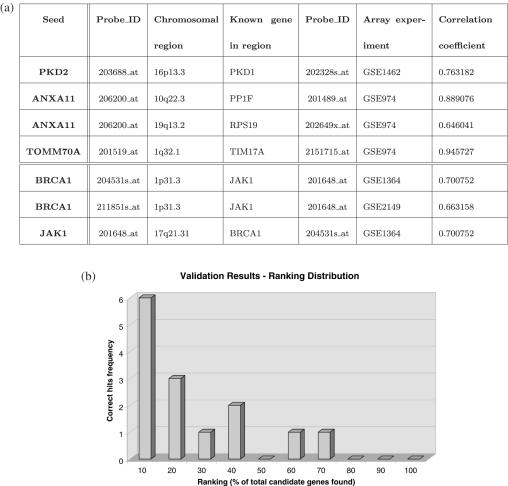
The following section presents results that show the ability of TOM to reproduce known genetic information (validation). We present three carefully documented benchmark tests whose results are summarized in the table of Figure 3a, and then broaden the validation with the analysis of five more examples. Global results are summarized in the rank distribution of Figure 3b that shows how the expected results rank in majority of the candidate genes list extracted with TOM.
Figure 3.
(a) The table summarizes the results of the first three examples. In the first four lines we record the results for One Locus problems for known interacting proteins. The last three lines show One Locus results for BRCA1-JAK1. (b) It shows a rank distribution of the genes known to be related to the eight examples discussed in Validation section, adding to the three described above five more benchmark examples, notably: Tuberous Sclerosis, Fanconi Anemia, Muscular Dystrophy, Myeloproliferative disorders and Neurotransmitter transport. The expected genes rank in majority within the first 20% of the list of candidate genes identified by TOM.
The Discovery section shows the results obtained on a Two Loci problem to gain further insight into a poorly characterized disease, namely familial thyroid cancer (discovery).
Validation
TOM was tested by searching for several genes known to interact with each other using the One Locus option of TOM. The aim of this approach was to ensure that the system correctly identifies gene–gene interplay. The examples used are reported in Figure 3a. Each gene was used as seed against the chromosomal region where the known interacting gene maps (ENSEMBL v.35), and vice versa. The examples considered for this first run of One Locus option were PKD1 and PKD2, T0MM70A and TIMM17A, ANNXA11 and PP1F.
PKD1 and PKD2 are genes mutated in polycystic kidney disease. In a majority of cases, the gene involved is PKD1, which is located on chromosome 16 (16q13.3) and encodes polycystin-1, a large receptor-like integral membrane protein. In the remaining (10–15%) cases, the disease is caused by mutational changes in another gene (PKD2), which is located at chromosome 4 (4q21–23) and encodes polcystin-2, a transmembrane protein, which acts as a non-specific calcium-permeable channel. Both polycystins function together in a non-redundant fashion, through a common pathway, and produce cellular responses that regulate proliferation, migration, differentiation and kidney morphogenesis [for a review see Ref. (23)].
TOMM70A and TIM17A are part of the mitochondrial complexes, through the outer and inner membrane respectively, for the import inside the mitochondria of nuclear-encoded proteins (21). Annexin 11 (ANXA11) is member of the annexin family, Ca2+-binding, membrane-fusogenic proteins with diverse functions. PP1F and RPS19 are two known interacting proteins with Annexin. Annexin 11 during cell cycle progression translocates from the nucleus to the spindle poles in metaphase and to the spindle midzone in anaphase (25).
We also tested the program for complex traits such as tumor predisposition and development. Since several genes are already known to be involved in predisposition to tumor, we tested TOM for the major gene for familial breast cancer, BRCA1. Loss of function of BRCA1 caused by inherited mutation and tissue-specific somatic mutation leads to breast and ovarian cancer. Nearly all BRCA1 germline mutations involve truncation or loss of the C-terminal BRCT transcriptional activation domain, suggesting that transcriptional regulation is a critical function of the wild-type gene. Several microarray analyses have been carried out to identify a peculiar gene expression profile characteristic of carriers of BRCA1 mutations, which would have an important impact also for diagnostic purpose [for an example see (26)]. It has been shown that there is a link between the role of BRCA1 in transcriptional regulation and its role in tumor suppression. Previous microarray analyses comparing transcription profiles of epithelial cells with low endogenous levels of BRCA1 versus transcription profiles of cells with 2 to 4-fold higher induced levels of expression of BRCA1 identified several genes with at least a 2-fold increase in expression, such as JAK1, a tyrosine protein kinase with a key role in cytokine signal transduction pathway (27). We thus tested whether by using BRCA1 as seed we could identify JAK1, giving as chromosomal location chr1p13.3, the region where JAK1 maps. The interaction was correctly identified, and also the reciprocal, i.e. JAK1 as seed and the region 17q21.31, where BRCA1 maps.
We also evaluated Tuberous Sclerosis with TSC1 and TSC2 involved genes, Fanconi Anemia with FANCA, FANCG and FANCL genes, Muscular Dystrophy with CAV3, CAPN3, TRIM32, SGCB, SGCG and DYSF genes, Myeloproliferative disorders with DTL and ZNF198, and finally the Neurotransmitter transport with NAT1 and NET1. We evaluated the correlations setting the threshold for FDR = 0.01. For these and previous results we ranked the candidate genes by correlation values (preserving the absolute values) using TOM automatic sorting of candidates genes based on correlation or corrected P-values. The ranking distribution is shown in Figure 3b.
Discovery—thyroid cancer
The TOM resource analyzes at the same time two different regions of interest and identifies the genes that are highly correlated and map to both regions. This approach proves very useful for genetic disorders in which a single gene has not yet been identified but genome scans provided regions of association on different chromosomes. We could hypothesize that genes with similar behavior might have a complementary effect on disease development.
We tested our hypothesis on the familial form of non-medullary thyroid carcinoma. Papillary thyroid carcinoma and follicular thyroid carcinoma are the most common forms of thyroid cancer accounting for between 80 and 90% of thyroid cancer patients. This disorder is associated with some of the highest familial risks among all cancer sites, with reported risks to first-degree relatives between 5- and 10-fold. Consequently, familial non-medullary thyroid cancer (fNMTC) has been recognized as a distinct clinicalentity, characterized by a higher degree of aggressiveness and mortality with respect to its sporadic counterpart (28). Transmission of susceptibility for fNMTC is compatible with an autosomal dominant mode of inheritance and incomplete penetrance. In collaboration with the International Consortium for the Genetics of fNMTC, two predisposing loci were previously mapped. The first one, TCO (Thyroid tumor with Cell Oxyphilia, MIM#603386), was mapped to the 19p13.2 region (29) and confirmed in additional families. Oxiphilic thyroid tumors are a particular form of thyroid neoplasia, characterized by cells with mitochondrial proliferation and hyperplasia, (oxyphilic or Huerthle cells). The second locus, NMTC1 (non-medullary thyroid carcinoma1), was mapped to chr2q21 and was associated with the follicular variant of PTC (fvPTC-MIM# 606240) (30). Evidence for an interaction between the two loci has been provided in a subset of fNMTC, and a two-locus mode of inheritance is consistent with stratification based on both the histological variants of oxyphilia and fvPTC (31).
We thus performed a search using TOM to verify whether the genes mapping to the two areas of interest have any degree of correlation between them, and they might also be considered as potential candidate genes based on their functions. Interestingly, 38 hits were identified and several genes showed a highly significant correlation and P-values (see Figure 2a and b). Among these genes, some look promising candidates for their biological function, such as UQCRFS1 on chromosome 19q, which is a mitochondrial ubiquitinol cytochrome c reductase iron–sulphur subunit and correlates with RAB3GAP on chromosome 2q, a Rab3 GTPase-protein involved in cell proliferation (correlation 0.626181; P-value < 0.001). This is very interesting since there is evidence of interaction between the two loci and the locus on chromosome 19 is associated with a mitochondrial phenotype. Thus, these genes could be considered plausible candidate genes based on position and function. Experimental studies will be needed to assess the presence of mutation/variants in affected individuals and prove an involvement in thyroid carcinoma predisposition. The advantage of using TOM here was to reduce the number of genes that can be selected for a first mutation screening, after having identified two regions of significant linkage.
CONCLUSIONS
We devised and implemented TOM, an algorithm for the identification of candidate genes responsible for genetic diseases. We took advantage of the microarray datasets available online to exploit novel computational biology approaches to molecular genetics. TOM allows a user to seamlessly associate functional and mapping data and to efficiently employ them in a quest for novel candidate genes in hereditary diseases.
Additional selection principles can be implemented to extend TOM, such as declaring a putative pathway for the candidate gene, in the case of poorly characterized diseases. Moreover, constant updating TOM with new expression datasets will increase the robustness of the assay. Our work represents a novel computational tool for gene hunters and could help to integrate and improve the comprehension of the genetic roots and the molecular mechanisms of complex life-threatening diseases.
Acknowledgments
The work was supported by grants from MIUR/PRIN and Telethon. The Open Access publication charge for this article has been waived by Oxford University Press.
Conflict of interest statement. None declared.
REFERENCES
- 1.The International HapMap Consortium. A haplotype map of the human genome. Nature. 2005;437:1299–1320. doi: 10.1038/nature04226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.ENCODE Consortium. The ENCODE (ENCylopedia Of DNA Elements) Project. Science. 2004;5696:636–640. doi: 10.1126/science.1105136. [DOI] [PubMed] [Google Scholar]
- 3.Hamosh A., Scott A.F., Amberger J., Bocchini C., Valle D., McKusick V.A. Online Mendelian Inheritance in Man (OMIM), a knowledgebase of human genes and genetic disorders. Nucleic Acids Res. 2002;30:52–55. doi: 10.1093/nar/30.1.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lapointe J., Li C., Higgins J.P., van de Rijn M., Bair E., Montgomery K., Ferrari M., Egevad L., Rayford W., Bergerheim U., et al. Gene expression profiling identifies clinically relevant subtypes of prostate cancer. Proc. Natl Acad. Sci. USA. 2004;101:811–816. doi: 10.1073/pnas.0304146101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liang Y., Diehn M., Watson N., Bollen A.W., Aldape K.D., Nicholas M.K., Lamborn K.R., Berger M.S., Botstein D., Brown P.O., Israel M.A. Gene expression profiling reveals molecularly and clinically distinct subtypes of glioblastoma multiforme. Proc. Natl Acad. Sci. USA. 2005;102:5814–5819. doi: 10.1073/pnas.0402870102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ramaswamy S., Ross K.N., Lander E.S., Golub T.R. A molecular signature of metastasis in primary solid tumors. Nature Genet. 2003;33:49–54. doi: 10.1038/ng1060. [DOI] [PubMed] [Google Scholar]
- 7.Sørlie T., Perou C.M., Tibshirani R., Aas T., Geisler S., Johnsen H., Hastie T., Eisen M.B., van de Rijn M., Jeffrey S., et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc. Natl Acad. Sci. 2001;98:10869–10874. doi: 10.1073/pnas.191367098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.The Gene Ontology Consortium. Creating the Gene Ontology resource: design and implementation. Genome Res. 2001;11:1425–1433. doi: 10.1101/gr.180801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Perez-Iratxeta C., Wjst M., Bork P., Andrade M.A. G2D: a tool for mining genes associated with disease. BMC Genet. 2005;6:45. doi: 10.1186/1471-2156-6-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Freudenberg J., Propping P. A similarity-based method for genome-wide prediction of disease-relevant human genes. Bioinformatics. 2002;18:S110–S115. doi: 10.1093/bioinformatics/18.suppl_2.s110. [DOI] [PubMed] [Google Scholar]
- 11.Adie E.A., Adams R.R., Evans K.L., Porteous D.J., Pickard B.S. Speeding disease gene discovery by sequence based candidate prioritization. BMC Bioinformatics. 2005;6:773–774. doi: 10.1186/1471-2105-6-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lopez-Bigas N., Ouzounis C.A. Genome-wide identification of genes likely to be involved in human genetic disease. Nucleic Acids Res. 2004;32:3108–3114. doi: 10.1093/nar/gkh605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.van Driel M.A., Cuelenaere K., Kemmeren P.P.C.W., Leunissen J.A.M., Brunner H.G., Vriend G. GeneSeeker: extraction and integration of human disease-related information from web-based genetic databases. Nucleic Acids Res. 2005;33:W758–W761. doi: 10.1093/nar/gki435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tiffin N., Kelso J.F., Powell A.R., Pan H., Bajic V.B., Hide W.A. Integration of text- and data-mining using ontologies successfully selects disease gene candidates. Nucleic Acids Res. 2005;33:1544–1552. doi: 10.1093/nar/gki296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Turner F.S., Clutterbuck D.R., Semple C.A. POCUS: mining genomic sequence annotation to predict disease genes. Genome Biol. 2003;4:R75. doi: 10.1186/gb-2003-4-11-r75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Apweiler R., Attwood T.K., Bairoch A., Bateman A., Birney E., Biswas M., Bucher P., Cerutti L., Corpet F., Croning M.D.R., et al. Interproan integrated documentation resource for protein families, domains and functional sites. Bioinformatics. 2000;16:1145–1150. doi: 10.1093/bioinformatics/16.12.1145. [DOI] [PubMed] [Google Scholar]
- 17.Franke L., van Bakel H., Fokkens L., de Jong E.D., Petersen M.-E., Wijmenga C. Reconstruction of functional human gene network, with an application for prioritizing positional candidate genes. Am. J. Hum. Genet. 2006;78:1011–1025. doi: 10.1086/504300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stelzl U., Worm U., Lalowski M., Haenig C., Brembeck F.H., Goehler H., Stroedicke M., Zenkner M., Schoenherr A., Koeppen S., et al. A human protein–protein interaction network: a resource for annotating the proteome. Cell. 2005;122:957–968. doi: 10.1016/j.cell.2005.08.029. [DOI] [PubMed] [Google Scholar]
- 19.Mootha V.K., Lepage P., Miller K., Bunkenborg J., Reich M., Hjerrild M., Delmonte T., Villeneuve A., Sladek R., Xu F., et al. Identification of a gene causing human cytochrome c oxidase deficiency by integrative genomics. Proc. Natl Acad. Sci. USA. 2003;100:605–610. doi: 10.1073/pnas.242716699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rosner B. Fundamentals of Biostatistics. Pacific Grove, CA: Duxbury; 2000. [Google Scholar]
- 21.Benjamini Y., Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. B. 1995;57:289–300. [Google Scholar]
- 22.Gentleman R.C., Carey V.J., Bates D.M., Bolstad B., Dettling M., Dudoit S., Ellis B., Gautier L., Ge Y., Gentry J., et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Al-Bhalal L., Akhtar M. Molecular basis of autosomal dominant polycystic kidney disease. Adv. Anat. Pathol. 2005;12:126–133. doi: 10.1097/01.pap.0000163959.29032.1f. [DOI] [PubMed] [Google Scholar]
- 24.Rapaport D. How does the TOM complex mediate insertion of precursor proteins into the mitochondrial outer membrane? J. Cell Biol. 2005;171:419–423. doi: 10.1083/jcb.200507147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tomas A., Futter C., Moss S.E. Annexin 11 is required for midbody formation and completion of the terminal phase of cytokinesis. J. Cell Biol. 2004;165:813–822. doi: 10.1083/jcb.200311054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.van't Veer L.J., Dai H., van de Vijver M.J., He Y.D., Hart A.A., Mao M., Peterse H.L., van der Kooy K., Marton M.J., Witteveen A.T., et al. Gene expression profiling predicts clinical outcome of breast cancer. Nature. 2002;415:530–536. doi: 10.1038/415530a. [DOI] [PubMed] [Google Scholar]
- 27.Welcsh P.L., Lee M.K., Gonzalez-Hernandez R.M., Black D.J., Mahadevappa M., Swisher E.M., Warrington J.A., King M.C. BRCA1 transcriptionally regulates genes involved in breast tumorigenesis. Proc. Natl Acad. Sci. USA. 2002;99:7560–7565. doi: 10.1073/pnas.062181799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Alsanea O., Clark O.H. Familial thyroid cancer. Curr. Opin. Oncol. 2001;13:44–51. doi: 10.1097/00001622-200101000-00009. [DOI] [PubMed] [Google Scholar]
- 29.Canzian F., Amati P., Harach H.R., Kraimps J.L., Lesueur F., Barbier J., Levillain P., Romeo G., Bonneau D. A gene predisposing to familial thyroid tumors with cell oxyphilia maps to chromosome 19p13.2. Am. J. Hum. Genet. 1998;63:1743–1748. doi: 10.1086/302164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McKay J.D., Lesueur F., Jonard L., Pastore A., Williamson J., Hoffman L., Burgess J., Duffield A., Papotti M., Stark M., et al. Localization of a susceptibility gene for familial nonmedullary thyroid carcinoma to chromosome 2q21. Am. J. Hum. Genet. 2001;69:440–446. doi: 10.1086/321979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McKay J.D., Thompson D., Lesueur F., Stankov K., Pastore A., Watfah C., Strolz S., Riccabona G., Moncayo R., Romeo G., Goldgar D.E. Evidence for interaction between the TCO and NMTC1 loci in familial non-medullary thyroid cancer. J. Med. Genet. 2004;41:407–412. doi: 10.1136/jmg.2003.017350. [DOI] [PMC free article] [PubMed] [Google Scholar]