Abstract
We have developed a web tool, PupaSuite, for the selection of single nucleotide polymorphisms (SNPs) with potential phenotypic effect, specifically oriented to help in the design of large-scale genotyping projects. PupaSuite uses a collection of data on SNPs from heterogeneous sources and a large number of pre-calculated predictions to offer a flexible and intuitive interface for selecting an optimal set of SNPs. It improves the functionality of PupaSNP and PupasView programs and implements new facilities such as the analysis of user's data to derive haplotypes with functional information. A new estimator of putative effect of polymorphisms has been included that uses evolutionary information. Also SNPeffect database predictions have been included. The PupaSuite web interface is accessible through http://pupasuite.bioinfo.cipf.es and through http://www.pupasnp.org.
INTRODUCTION
Single nucleotide polymorphisms (SNPs) are the simplest and most frequent type of DNA sequence variation among individuals and constitute one of the most powerful tools in the search for disease susceptibility genes, drug response-determining genes and the like (1,2). With the introduction of large-scale genotyping techniques the bottleneck in this type of experiments has moved towards the management and analysis of the data generated. In this context, one of the topics which has become a problem is the step of the selection of the optimal set of SNPs (among several thousands of candidates in some cases) for the genotyping experiment. Optimal SNPs must be the best possible markers for traits, which often are multigenic, usually reflecting disruptions in proteins that participate in a protein complex or a in a pathway (3). Unfortunately, complex multigenic traits, for which markers display weak associations, still constitute a challenge. Factors such as linkage disequilibrium (LD) and minor allele frequency (MAF) are of major importance for selecting optimal candidate SNPs. Recently, the predicted functional effect of an SNP is gaining importance as a selection criterium because it constitutes a potential important factor for increasing the sensitivity of association tests significantly (3–6). The availability of information on LD from projects such as HapMap (7), on MAFs (8) and improved methods for predicting function (5,6,9), allow for a more sophisticated selection of candidate SNPs beyond the classical one-SNP-at-a-time approach. Thus, SNPs can be selected taking into account the evolutionary constraints of the region analysed along with its likelihood of being the causative agent of any type of damage. Algorithms which use information to facilitate the posterior analysis of the results, such as the estimation of haplotype blocks (10), combined with functional prediction of the effect of the SNPs, are expected to have a major impact on the efficiency of a large-scale genotyping study. PupaSuite belongs to this new generation of tools. PupaSuite combines the facilities offered by PupaSNP (6) and PupasView (5) with new algorithms and visualisation procedures for functional haplotype prediction. The PupaSNP and PupasView programs are part of the pipeline of genotyping of the Spanish National Genotyping Center (CeGen; http://www.cegen.org/). Both tools combined bear an average of 60 SNP designs per day.
OUTLINE OF THE PROGRAM
PupaSuite combines the functionality of PupaSNP (6) and PupasView (5) in a unique and more integrated interface, and adds new modules to facilitate the selection of the optimal set of SNPs for a large-scale genotyping study. Following the philosophy of PupaSNP, the program allows to input either lists of genes or chromosomal regions, which would correspond to two common types of analysis: genes probably related to a disease because they are functionally related (e.g. they belong to a pathway affected in the disease), or genes present in a chromosomal region linked to a disease. PupaSuite can also directly analyse lists of SNPs. In these three cases a list of SNPs with their putative functional effect is reported. In the case of chromosomal regions it is also possible to find haplotype blocks (10). For the list of SNPs, in addition to their putative functional effect, it is possible to retrieve information on MAF in different populations from dbSNP (8) [as annotated in the Ensembl (11)], as well as LD parameters and haplotype blocks.
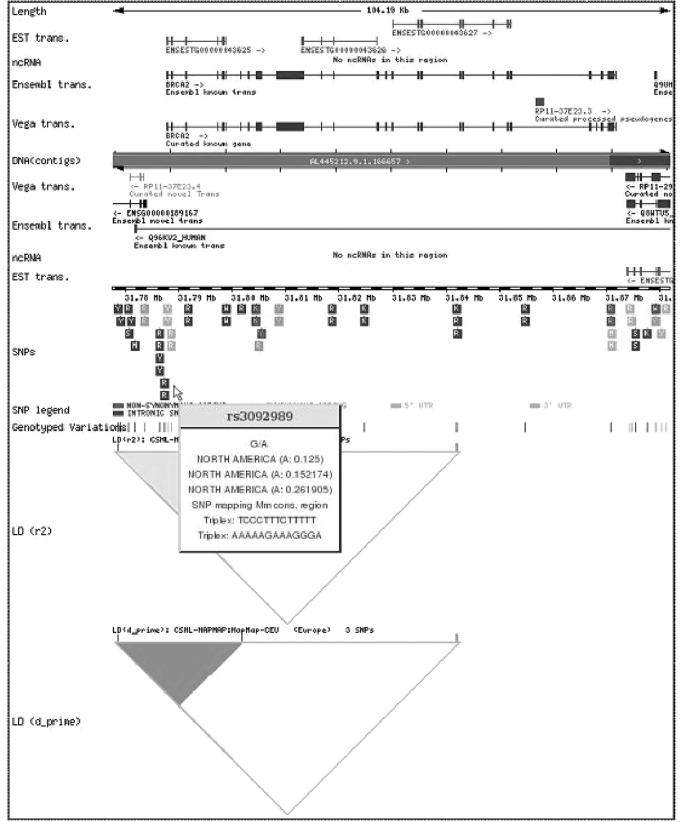
In addition to the analysis of lists of SNPs there is another new option: Functional haplotypes. This option (see below) allows the user to test their own SNP data and to find haplotypes (12) with the functional SNPs (5,6) and the tag SNPs (13) highlighted. Case-control studies can also be performed at this stage. The option Display and Filter SNPs for a single gene implements new functionalities in an environment a la PupasView (5). More information is presented in a graphical intuitive format (Figure 1). This option allows the sequential and interactive application of filters based on functionality, conservation, MAF and the like (5) thus permitting an easy selection of a set of optimal SNPs for a particular gene.
Figure 1.
Output with the graphic representation of SNPs with putative functional effect in the gene BRCA2, along with LD maps.
CRITERIA TO SELECT SNPS AS A GOOD CANDIDATES FOR GENOTYPING
Here three important features of a SNP have been taken into account in order to be considered as an optimal candidate for genotyping purposes: MAF, LD with respect to other candidates (5) and putative functional effect. MAF values were taken from the Ensembl (11), which maps dbSNP (8) data onto the corresponding chromosomal coordinates. LD are calculated as r2 and D′ with the Haploview program (14). The putative functional effect has been estimated in both coding and non-coding regions as described in (5). The following features have been used to report the putative functional effect of a polymorphism in non-coding nucleotides:
Transcription factor binding sites from the Transfac database (15).
Intron/exon border consensus sequences.
Exonic splicing enhancers (16).
Triplex-forming oligonucleotide target sequences (17).
Regarding the putative impact of a cSNP, the following data and estimators are reported:
SNPs in exons causing an amino acid change (purely a list of cSNPs)
Selective strengths (ω parameter). This estimator is new in this version of the program (see below)
SNPeffect (9,20,21) predictions. New in this version of the program (see below).
The likelihood of the predictions can be reinforced by looking simultaneously for human-mouse conserved regions (22) as reported in Ensembl (http://www.ensembl.org).
EVOLUTION AT WORK: THE SELECTIVE STRENGTHS ON CSNPS
The combined effect of all the selective pressures causes the preservation of the functionally relevant parts of the genes. Under this perspective, comparative and evolutionary studies have been used to predict the putative functional effect of SNPs (19,23) although these have mainly ignored the underlying phylogeny. Here we present another more accurate estimator of functional effect, based on sequence comparison, but taking into account phylogenetic information (24). The selective pressures acting at a codon-level where non-synonymous cSNPs are found were evaluated by means of two alternative approaches: codon-based maximum likelihood (ML) models (25) implemented in PAML (26), and likelihood-ratio (SLR) method (27) for testing deviations of neutrality.
Under the first approximation, an a priori statistical distribution describing the variation of ω = dN/dS among sites is assumed for a number k of different classes of sites with ωk values at a proportion pk of the sequences representing the effects of purifying selection (0 < ω0 < 1), neutral evolution (ω1 = 1), and positive selection (ω2 > 1) (25). The method involves two main steps: first, the adjustment by maximum likelihood of the evolutionary parameters to the sequences of the species compared considering two different models; and second, the use of the Bayes theorem to compute the posterior probability that each site belongs to a specific site class ωk defined under an a priori distribution (28). Two different models (M2a and M8) were evaluated by maximum likelihood on the sequences (29).
Under the sitewise likelihood-ratio method (SLR) a site-by-site approach to test for neutrality is used. In contrast to similar approaches developed previously (30), SLR uses the entire alignment of the sequence to determine parameters common to all sites, such as evolutionary distances. Using this approach there is no need to specify a model of how ω varies along the sequence. A correction for multiple testing in order to obtain statistical confidence for inferences on deviations from neutrality on each site is also performed.
SNPEFFECT DATABASE
The SNPeffect database (9) describes the effect of coding non-synonymous SNPs on several phenotypic properties of human proteins using either sequence-based or structural bioinformatics tools. Molecular phenotypes are grouped in three categories: structure and dynamics, functional sites and cellular processing. Next to various external tools SNPeffect uses algorithms developed at the collaborating research groups, among which Tango (20) to predict β-aggregation regions in protein sequences and FoldX (21) to predict the stability change caused by the single amino acid variation.
FUNCTIONAL HAPLOTYPES
In addition to using already available data, the users can input their own data to use the predictions on possible functional effects in combination with haplotype analysis. This possibility can be used through the Functional haplotypes option. Data must be provided to the program in linkage pedigree format (pre MAKEPRED, http://pupasuite.bioinfo.cipf.es/html/help/index.html). The PupaSuite estimates blocks by three methods: Confidence intervals (10), Four gamete rule (31) and Solid Spine of LD (14) and reconstruct haplotypes using the EM algorithm (12) as implemented in Haploview (14). The haplotypes found in this way are represented with the corresponding functional information on all the SNPs included in it and all the LD values. This representation provides a very intuitive picture of the possible functional impact of any of the haplotypes beyond the individual effect of each SNP. For case/control data a chi-square test is performed and the corresponding P-value for the allele frequencies in cases versus control is reported. The combination of functional haplotype information with case/control tests allows to easily ascribe cases to haplotypes with functional alterations.
DISCUSSION
We have presented an integrated resource for helping in the selection of optimal sets of SNPs oriented to large-scale genotyping assays. The program merges the functionalities of other two previous resources, PupaSNP (6) and PupasView (5), and expand the capabilities of the program with new information and new facilities. The SNPeffect database (9) as well as a new, unpublished prediction method has been included to improve the estimation of the putative pathological effect of SNPs. Moreover, in addition to use publicly available data on SNPs, users can analyse their own experiments. What is novel and unique to tools of this type is the possibility of analysing functionally haplotypes, beyond the classical analysis one-SNP-at-a-time which ignores interactions between the mutations.
The usefulness of this type of resources is proven by the use made by the CeGen in its pipeline of genotyping. The previous tools, which have been running for more than two years, have now an approximate average of 60 daily SNP designs (http://bioinfo.cipf.es/webalizer/pupasnp and http://bioinfo.cipf.es/webalizer/pupasview).
Acknowledgments
This work is supported by grants from Fundació La Caixa, Fundación BBVA, MEC BIO2005-01078 and NRC Canada-SEPOCT Spain. The Functional Genomics node (INB) is supported by Genoma España. LC is supported by fellowship from the CeGen (Genoma España). Funding to pay the Open Access publication charges for this article was provided by Genome España.
Conflict of interest statement. None declared.
REFERENCES
- 1.Collins F.S., Green E.D., Guttmacher A.E., Guyer M.S. A vision for the future of genomics research. Nature. 2003;422:835–847. doi: 10.1038/nature01626. [DOI] [PubMed] [Google Scholar]
- 2.Risch N.J. Searching for genetic determinants in the new millennium. Nature. 2000;405:847–856. doi: 10.1038/35015718. [DOI] [PubMed] [Google Scholar]
- 3.Badano J.L., Katsanis N. Beyond Mendel: an evolving view of human genetic disease transmission. Nature Rev. Genet. 2002;3:779–789. doi: 10.1038/nrg910. [DOI] [PubMed] [Google Scholar]
- 4.Botstein D., Risch N. Discovering genotypes underlying human phenotypes: past successes for mendelian disease, future approaches for complex disease. Nature Genet. 2003;33:228–237. doi: 10.1038/ng1090. [DOI] [PubMed] [Google Scholar]
- 5.Conde L., Vaquerizas J.M., Ferrer-Costa C., de la Cruz X., Orozco M., Dopazo J. PupasView: a visual tool for selecting suitable SNPs, with putative pathological effect in genes, for genotyping purposes. Nucleic Acids Res. 2005;33:W501–W505. doi: 10.1093/nar/gki476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Conde L., Vaquerizas J.M., Santoyo J., Al-Shahrour F., Ruiz-Llorente S., Robledo M., Dopazo J. PupaSNP Finder: a web tool for finding SNPs with putative effect at transcriptional level. Nucleic Acids Res. 2004;32:W242–W248. doi: 10.1093/nar/gkh438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Altshuler D., Brooks L.D., Chakravarti A., Collins F.S., Daly M.J., Donnelly P. A haplotype map of the human genome. Nature. 2005;437:1299–1320. doi: 10.1038/nature04226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wheeler D.L., Barrett T., Benson D.A., Bryant S.H., Canese K., Chetvernin V., Church D.M., DiCuccio M., Edgar R., Federhen S., et al. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2006;34:D173–D180. doi: 10.1093/nar/gkj158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Reumers J., Schymkowitz J., Ferkinghoff-Borg J., Stricher F., Serrano L., Rousseau F. SNPeffect: a database mapping molecular phenotypic effects of human non-synonymous coding SNPs. Nucleic Acids Res. 2005;33:D527–D532. doi: 10.1093/nar/gki086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gabriel S.B., Schaffner S.F., Nguyen H., Moore J.M., Roy J., Blumenstiel B., Higgins J., DeFelice M., Lochner A., Faggart M., et al. The structure of haplotype blocks in the human genome. Science. 2002;296:2225–2229. doi: 10.1126/science.1069424. [DOI] [PubMed] [Google Scholar]
- 11.Hubbard T., Andrews D., Caccamo M., Cameron G., Chen Y., Clamp M., Clarke L., Coates G., Cox T., Cunningham F., et al. Ensembl 2005. Nucleic Acids Res. 2005;33:D447–D453. doi: 10.1093/nar/gki138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Qin Z.S., Niu T., Liu J.S. Partition-ligation-expectation-maximization algorithm for haplotype inference with single-nucleotide polymorphisms. Am. J. Hum. Genet. 2002;71:1242–1247. doi: 10.1086/344207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.de Bakker P.I., Yelensky R., Pe'er I., Gabriel S.B., Daly M.J., Altshuler D. Efficiency and power in genetic association studies. Nature Genet. 2005;37:1217–1223. doi: 10.1038/ng1669. [DOI] [PubMed] [Google Scholar]
- 14.Barrett J.C., Fry B., Maller J., Daly M.J. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 15.Wingender E., Chen X., Hehl R., Karas H., Liebich I., Matys V., Meinhardt T., Pruss M., Reuter I., Schacherer F. TRANSFAC: an integrated system for gene expression regulation. Nucleic Acids Res. 2000;28:316–319. doi: 10.1093/nar/28.1.316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cartegni L., Chew S.L., Krainer A.R. Listening to silence and understanding nonsense: exonic mutations that affect splicing. Nature Rev. Genet. 2002;3:285–298. doi: 10.1038/nrg775. [DOI] [PubMed] [Google Scholar]
- 17.Goni J.R., de la Cruz X., Orozco M. Triplex-forming oligonucleotide target sequences in the human genome. Nucleic Acids Res. 2004;32:354–360. doi: 10.1093/nar/gkh188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ferrer-Costa C., Orozco M., de la Cruz X. Characterization of disease-associated single amino acid polymorphisms in terms of sequence and structure properties. J. Mol. Biol. 2002;315:771–786. doi: 10.1006/jmbi.2001.5255. [DOI] [PubMed] [Google Scholar]
- 19.Ferrer-Costa C., Orozco M., de la Cruz X. Sequence-based prediction of pathological mutations. Proteins. 2004;57:811–819. doi: 10.1002/prot.20252. [DOI] [PubMed] [Google Scholar]
- 20.Fernandez-Escamilla A.M., Rousseau F., Schymkowitz J., Serrano L. Prediction of sequence-dependent and mutational effects on the aggregation of peptides and proteins. Nat. Biotechnol. 2004;22:1302–1306. doi: 10.1038/nbt1012. [DOI] [PubMed] [Google Scholar]
- 21.Schymkowitz J., Borg J., Stricher F., Nys R., Rousseau F., Serrano L. The FoldX web server: an online force field. Nucleic Acids Res. 2005;33:W382–W388. doi: 10.1093/nar/gki387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schwartz S., Kent W.J., Smit A., Zhang Z., Baertsch R., Hardison R.C., Haussler D., Miller W. Human-mouse alignments with BLASTZ. Genome Res. 2003;13:103–107. doi: 10.1101/gr.809403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Miller M.P., Kumar S. Understanding human disease mutations through the use of interspecific genetic variation. Hum. Mol. Genet. 2001;10:2319–2328. doi: 10.1093/hmg/10.21.2319. [DOI] [PubMed] [Google Scholar]
- 24.Arbiza L., Duchi S., Montaner D., Burguet J., Pantoja-Uceda D., Pineda-Lucena A., Dopazo J., Dopazo H. Selective pressures at a codon-level predict deleterious mutations in human disease genes. J. Mol. Biol. 2006 doi: 10.1016/j.jmb.2006.02.067. in press. [DOI] [PubMed] [Google Scholar]
- 25.Yang Z., Nielsen R. Codon-substitution models for detecting molecular adaptation at individual sites along specific lineages. Mol. Biol. Evol. 2002;19:908–917. doi: 10.1093/oxfordjournals.molbev.a004148. [DOI] [PubMed] [Google Scholar]
- 26.Yang Z. PAML: a program package for phylogenetic analysis by maximum likelihood. Comput. Appl. Biosci. 1997;13:555–556. doi: 10.1093/bioinformatics/13.5.555. [DOI] [PubMed] [Google Scholar]
- 27.Massingham T., Goldman N. Detecting amino acid sites under positive selection and purifying selection. Genetics. 2005;169:1753–1762. doi: 10.1534/genetics.104.032144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yang Z., Wong W.S., Nielsen R. Bayes empirical bayes inference of amino acid sites under positive selection. Mol. Biol. Evol. 2005;22:1107–1118. doi: 10.1093/molbev/msi097. [DOI] [PubMed] [Google Scholar]
- 29.Yang Z., Nielsen R., Goldman N., Pedersen A.M. Codon-substitution models for heterogeneous selection pressure at amino acid sites. Genetics. 2000;155:431–449. doi: 10.1093/genetics/155.1.431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Suzuki Y., Gojobori T. A method for detecting positive selection at single amino acid sites. Mol. Biol. Evol. 1999;16:1315–1328. doi: 10.1093/oxfordjournals.molbev.a026042. [DOI] [PubMed] [Google Scholar]
- 31.Wang N., Akey J.M., Zhang K., Chakraborty R., Jin L. Distribution of recombination crossovers and the origin of haplotype blocks: the interplay of population history, recombination, and mutation. Am. J Hum. Genet. 2002;71:1227–1234. doi: 10.1086/344398. [DOI] [PMC free article] [PubMed] [Google Scholar]