Abstract
Identification of MHC-binding peptides is a prerequisite in rational design of T-cell based peptide vaccines. During the past decade a number of computational approaches have been introduced for the prediction of MHC-binding peptides, efficiently reducing the number of candidate binders that need to be experimentally verified. Here the SVMHC server for prediction of both MHC class I and class II binding peptides is presented. SVMHC offers fast analysis of a wide range of alleles and prediction results are given in several comprehensive formats. The server can be used to find the most likely binders in a protein sequence and to investigate the effects of single nucleotide polymorphisms in terms of MHC-peptide binding. The SVMHC server is accessible at http://www-bs.informatik.uni-tuebingen.de/SVMHC/.
INTRODUCTION
The immune system provides an effective line of defense against invading pathogens and cancer. The adaptive part of the immune system, which is responsible for specific recognition of antigen and immunological memory, is highly dependent on the activation of T-cells. T-cells only recognize antigenic peptides bound to major histocompatibility (MHC) molecules on the surface of other cells. This makes MHC-peptide binding a prerequisite for T-cell activation. There are two major classes of MHC molecules. MHC class I molecules typically bind peptides that are 9 amino acids long and originate from intracellular proteins. Intracellular proteins are continuously degraded into smaller peptides that are displayed on the cells surface by MHC molecules, giving a kind of fingerprint of the cellular proteome. This mechanism ensures that virally infected cells or cancer cells can be detected, since virus or cancer-specific MHC-peptide complexes are displayed on the cell surface. Cytotoxic T-cells (CD8+) of the immune system can recognize such abnormal cells and eliminate them. MHC class II molecules, on the other hand, bind peptides originating from extracellular antigens. These peptides are usually longer compared with MHC class I peptides (15–25 amino acids), however the main part of the MHC-peptide interactions is given by a binding core of 9 amino acids. MHC class II molecules are mainly presented on antigen presenting cells (APCs) and activate helper T-cells (CD4+). In recent years, MHC-binding peptides have proven useful for immunotherapeutic purposes in studies concerning both different cancer types (1,2) and HIV infection (3). The aim of these approaches is to use antigen-specific peptides in order to activate the immune system. The first step here is to find a set of MHC-binding peptides given an antigen of interest. One challenge here is the extreme variability of the MHC molecules with many hundred allelic variants. However, typically only one in 100–200 potential peptides actually binds to a certain MHC allele (4). This has motivated computational approaches for modeling MHC allele-specific peptide preferences. Such methods can reduce the number of peptides that have to be verified experimentally.
The first prediction methods utilized simple sequence motif searches for identifying potential MHC class I binding peptides (5,6). These methods have since been refined into position-specific scoring matrix (PSSM) approaches (7–13). One drawback of these methods is that they assume an independent contribution of each amino acid in the peptides to the overall binding affinity, neglecting the effects of neighbouring residues. An obvious case where this might be a problem, is when two compete for the same space in a binding pocket. Several machine learning methods have been introduced that aim to model the MHC-peptide interaction in a non-linear fashion (14–18), potentially overcoming the limitation of PSSM-based methods. The above mentioned methods are all sequence-based, but a number of structure-based methods have also been presented (19–22).
Prediction of MHC class II peptides is more challenging, owing to the additional alignment needed to identify the binding core within the longer peptides. Once the sequences have been correctly aligned, the computational problem is very similar to that of the class I case. Methods for MHC class II prediction include genetic algorithms coupled with neural networks (23) and Gibbs sampling (24), as well as the construction of PSSMs using virtual binding pockets (25). The predicted MHC class II binding cores are often extended at both ends to obtain an effective T-cell epitope.
Here, the SVMHC server for prediction of MHC class I and class II binding peptides is presented. In contrast to most other prediction servers, SVMHC offers several comprehensive results formats, easy access to data from protein databases and refinement of initial predictions. Furthermore, SVMHC enables analysis of the effects of single nucleotide polymorphisms (SNPs) in terms of MHC-peptide binding. A number of new prediction models for human and mouse MHC class I molecules have been added. Furthermore, MHC class II prediction can be done utilizing the matrices published by Sturniolo et al. (25).
THE SVMHC PREDICTION SERVER
Prediction models
A support vector machine (SVM) approach is used for the prediction of MHC class I binding peptides. This approach has been described in detail in a previous publication (17) and is only briefly outlined here. MHC-binding peptides of different lengths were extracted from the MHCPEP (26) and SYFPEITHI (8) databases. The main difference between these two data sources is that MHCPEP contains both naturally processed and synthetic peptides, whereas SYFPEITHI exclusively contains naturally processed peptides. In order to construct the prediction models, each peptide was represented using binary sparse encoding. Different kernels and a grid search strategy were then employed to find optimal SVM parameters. Approximately 20 known binders are needed in order to construct prediction models with significant accuracy. For most alleles prediction models could only be generated for peptides with a length of nine amino acids due to the amount of data available. However, in some cases prediction models were also constructed for peptides with a length of eight or ten amino acids. In comparative studies against the prediction methods BIMAS (9) and SYFPEITHI (8), SVMHC showed improved performance for most MHC alleles (17). Prediction models are now available for 26 different human MHC alleles based on data from MHCPEP. Prediction models based on data from SYFPEITHI are available for 19 human and 5 murine MHC alleles.
Prediction of MHC class II binding peptides is based on the matrices published by Sturniolo et al. (25). By sequence similarity studies, they defined modular pockets in the MHC molecule involved in peptide interaction. These pockets are independent of the rest of the binding cleft and a limited number of pockets can be combined into virtual binding matrices for a wide range of MHC class II alleles. These matrices are also a part of the TEPITOPE prediction software and they have been used to identify candidate binding peptides for both HIV (3) and Tuberculosis (27) vaccines. Prediction is available for 51 different MHC class II alleles.
Whole protein prediction
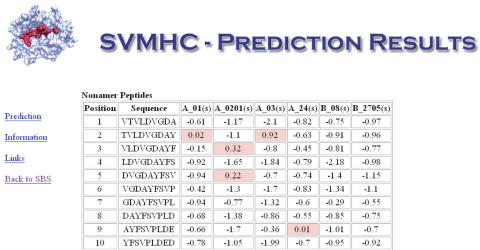
The input required for analysis by SVMHC is a protein sequence and a specification of one or more MHC alleles. The protein sequence can either be directly pasted into the web interface or accessed directly by entering a database ID from the NCBI RefSeq (28) or Swiss-Prot (29) databases. Prediction is carried out for all possible peptides of the protein using a sliding window. Several different output formats are given in order to facilitate further analysis. The default output format is a list of putative binders, where the best binder is found at the top. A summary table is also generated for all peptides of a certain length. The summary table shows the results ordered according to peptide start position in the protein (see Figure 1 for an example).
Figure 1.
A summary table produced by SVMHC showing peptide start position, sequence and allele-specific score. Predicted MHC binders are highlighted in red, enabling fast identification of peptides binding to several different MHC alleles.
Binders are highlighted, which enables fast identification of peptides likely to bind several MHC alleles, so called promiscuous epitopes. These are especially interesting for vaccine design since they cover a wider range of the population. A graphical overview is also given to further facilitate the identification of promiscuous epitopes (see Figure 2).
Figure 2.
A graphical view of the predicted MHC-binding peptides. Predicted binding peptides are colored red, except for the first amino acid that is colored blue. This view enables a fast scan, even of long proteins, in order to identify promiscuous epitopes and epitope-rich regions.
The initial prediction results can then be further refined by removing or adding alleles of interest. The complete prediction results can also be downloaded in tab-separated format, enabling further analysis in any spreadsheet-based program (e.g. Microsoft Excel).
Analyzing the effects of SNPs
Several studies have pointed out the importance of SNPs in terms of MHC-binding peptides (30–32). SVMHC allows for the analysis of SNPs in terms of MHC-peptide binding. For this analysis a protein sequence and a specified mutation (e.g. A23P, meaning that alanine in position 23 of the protein is changed to a proline) is required. All relevant peptides, with and without the mutation, are then generated and predicted by SVMHC. The results are presented in a comparative manner, highlighting the effects of the amino acid substitution. A good example for this type of analysis is the well-known HLA-A*03-restricted epitope RLRPGGKKK originating from the HIV matrix protein p17 (30,31). Studies have identified polymorphisms within this peptide, where the exchange of the lysine in position nine to a threonine, cause viral escape (31) (meaning that the mutated peptides are not recognized by the immune system). The whole p17 protein with the specified K27T mutation (corresponding to the mutation in position nine of the peptide of interest) was analyzed by SVMHC, see Figure 3.
Figure 3.
Prediction results for analyzing the effect of the K27T mutation in the HIV matrix protein p17. From these results it can be seen that the peptide binding is substantially reduced, a possible explanation for immune escape. Predicted binders are highlighted green and if the difference in the predicted score between two binders is <0.5, the score difference is highlighted blue.
The mutation substantially reduces the predicted MHC affinity of the peptide. SNPs can also influence amino acids of the peptide, which are less involved in MHC binding and rather important for T-cell recognition. Binders and non-binders are highlighted in green and red, respectively, in the result table (if at least one peptides is predicted as a binder). Furthermore, a blue highlighting is given if the difference between two peptides is >0.5. The dynamic coloring makes it easy to identify SNPs that are interesting for further analysis.
CONCLUSION
We present an updated and extended version of the SVMHC server for predicting MHC-binding epitopes. SVMHC combines high prediction accuracy with a wide range of both, MHC class I and MHC class II alleles. Compared with other prediction tools, it also provides a number of different output formats ranging from summary graphical views to detailed comparison tables. All data can also be exported for external analysis. Another singular feature of SVMHC is the ability to analyze the effects of SNPs on MHC epitopes. This type of analysis is interesting for viral epitopes (prediction of immune escape) and the analysis of minor histocompatibility antigens (miHAgs). SVMHC is updated regularly and as new MHC-binding data becomes available it will be integrated into SVMHC. This ensures continual improvement of both prediction accuracy and allele coverage.
Acknowledgments
This work was supported by the Deutsche Forschungsgemeinschaft (SFB 685). Funding to pay the Open Access publication charges for this article was provided by the Deutsche Forschungsgemeinschaft (SFB 685).
Conflict of interest statement. None declared.
REFERENCES
- 1.Hsu F.J., Benike C., Fagnoni F., Liles T.M., Czerwinski D., Taidi B., Engleman E.G., Levy R. Vaccination of patients with B-cell lymphoma using autologous antigen-pulsed dendritic cells. Nat. Med. 1996;2:52–58. doi: 10.1038/nm0196-52. [DOI] [PubMed] [Google Scholar]
- 2.Nestle F.O., Alijagic S., Gilliet M., Sun Y., Grabbe S., Dummer R., Burg G., Schadendorf D. Vaccination of melanoma patients with peptide- or tumor lysate-pulsed dendritic cells. Nat. Med. 1998;4:328–332. doi: 10.1038/nm0398-328. [DOI] [PubMed] [Google Scholar]
- 3.De Groot A.S., Marcon L., Bishop E.A., Rivera D., Kutzler M., Weiner D.B., Martin W. HIV vaccine development by computer assisted design: the GAIA vaccine. Vaccine. 2005;23:2136–2148. doi: 10.1016/j.vaccine.2005.01.097. [DOI] [PubMed] [Google Scholar]
- 4.Yewdell J.W., Bennink J.R. Mechanisms of viral interference with MHC class I antigen processing and presentation. Annu. Rev. Cell Dev. Biol. 1999;15:579–606. doi: 10.1146/annurev.cellbio.15.1.579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sette A., Buus S., Appella E., Smith J.A., Chesnut R., Miles C., Colon S.M., Grey H.M. Prediction of major histocompatibility complex binding regions of protein antigens by sequence pattern analysis. Proc. Natl Acad. Sci. USA. 1989;86:3296–3300. doi: 10.1073/pnas.86.9.3296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rötzschke O., Falk K., Stevanovic S., Jung G., Walden P., Rammensee H.G. Exact prediction of a natural T cell epitope. Eur. J. Immunol. 1991;21:2891–2894. doi: 10.1002/eji.1830211136. [DOI] [PubMed] [Google Scholar]
- 7.Kondo A., Sidney J., Southwood S., delGuercio M.F., Appella E., Sakamoto H., Celis E., Grey H.M., Chesnut R.W., Kubo R.T., et al. Prominent roles of secondary anchor residues in peptide binding to HLA-A24 human class I molecules. J. Immunol. 1995;155:4307–4312. [Google Scholar]
- 8.Rammensee H.-G., Bachman J., Philipp N., Emmerich N., Bachor O.A., Stevanovic S. SYFPEITHI: a database for MHC ligands and peptide motifs. Immunogenetics. 1997;50:213–219. doi: 10.1007/s002510050595. [DOI] [PubMed] [Google Scholar]
- 9.Parker K.C., Bednarek M.A., Coligan J.E. Scheme for ranking potential HLA-A2 binding peptides based on independent binding of individual peptide side-chains. J. Immunol. 1994;152:163–175. [PubMed] [Google Scholar]
- 10.Reche P.A., Glutting J.-P., Zhang H., Reinherz E.L. Enhancement to the RANKPEP resource for the prediction of peptide binding to MHC molecules using profiles. Immunogenetics. 2004;56:405–419. doi: 10.1007/s00251-004-0709-7. [DOI] [PubMed] [Google Scholar]
- 11.Sidney J., Grey H.M., Southwood S., Celis E., Wentworth P.A., delGuercio M.F., Kubo R.T., Chesnut R.W., Sette A. Definition of an HLA-A3-like supermotif demonstrates the overlapping peptide-binding repertoires of common HLA molecules. Hum. Immunol. 1996;45:79–93. doi: 10.1016/0198-8859(95)00173-5. [DOI] [PubMed] [Google Scholar]
- 12.Sidney J., Southwood S., delGuercio M.F., Grey H.M., Chesnut R.W., Kubo R.T., Sette A. Specificity and degeneracy in peptide binding to HLA-B7-like class I molecules. J. Immunol. 1996;157:3480–3490. [PubMed] [Google Scholar]
- 13.Sidney J., Southwood S., Pasquetto V., Sette A. Simultaneous prediction of binding capacity for multiple molecules of the HLA B44 supertype. J. Immunol. 2003;171:5964–5974. doi: 10.4049/jimmunol.171.11.5964. [DOI] [PubMed] [Google Scholar]
- 14.Gulukota K., Sidney J., Sette A., DeLisi C. Two complementary methods for predicting peptides binding major histocompatibility complex molecules. J. Mol. Biol. 1997;267:1258–1267. doi: 10.1006/jmbi.1997.0937. [DOI] [PubMed] [Google Scholar]
- 15.Honeyman M.C., Brusic V., Stone,N.L., Harrison L.C. Neural network-based prediction of candidate T-cell epitopes. Nat. Biotechnol. 1998;16:966–969. doi: 10.1038/nbt1098-966. [DOI] [PubMed] [Google Scholar]
- 16.Mamitsuka H. Predicting peptides that bind to MHC molecules using supervised learning of hidden markov models. Proteins. 1998;33:460–474. doi: 10.1002/(sici)1097-0134(19981201)33:4<460::aid-prot2>3.0.co;2-m. [DOI] [PubMed] [Google Scholar]
- 17.Dönnes P., Elofsson A. Prediction of MHC class I binding peptides, using SVMHC. BMC Bioinformatics. 2002;3:25. doi: 10.1186/1471-2105-3-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nielsen M., Lundegaard C., Worning P., Lauemoller S.L., Lamberth K., Buus S., Brunak S., Lund O. Reliable prediction of T-cell epitopes using neural networks with novel sequence representations. Protein Sci. 2003;12:1007–1017. doi: 10.1110/ps.0239403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rognan D., Scapozza L., Folkers G., Daser A. Molecular dynamics simulation of MHC-peptide complexes as a tool for predicting potential T cell epitopes. Biochemistry. 1994;33:11476–11485. doi: 10.1021/bi00204a009. [DOI] [PubMed] [Google Scholar]
- 20.Logean A., Rognan D. Recovery of known T-cell epitopes by computational scanning of a viral genome. J. Comput. Aided Mol. Des. 2002;16:229–243. doi: 10.1023/a:1020244329512. [DOI] [PubMed] [Google Scholar]
- 21.Tong J.C., Tan T.W., Ranganathan S. Modeling the structure of bound peptide ligands to major histocompatibility complex. Protein Sci. 2004;13:2523–2532. doi: 10.1110/ps.04631204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schueler-Furman O., Altuvia Y., Sette A. Structure-based prediction of binding peptides to MHC class I molecules: application to a broad range of MHC alleles. Protein Sci. 2000;9:1838–1846. doi: 10.1110/ps.9.9.1838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brusic V., Rudy G., Honeyman G., Hammer J., Harrison L. Prediction of MHC class II-binding peptides using an evolutionary algorithm and artificial neural network. Bioinformatics. 1998;14:121–130. doi: 10.1093/bioinformatics/14.2.121. [DOI] [PubMed] [Google Scholar]
- 24.Nielsen M., Lundegaard C., Worning P., Hvid C.S., Lamberth K., Buus S., Brunak S., Lund O. Improved prediction of MHC class I and class II epitopes using a novel Gibbs sampling approach. Bioinformatics. 2004;20:1388–1397. doi: 10.1093/bioinformatics/bth100. [DOI] [PubMed] [Google Scholar]
- 25.Sturniolo T., Bono E., Ding J., Raddrizzani L., Tuereci O., Sahin U., Braxenthaler M., Gallazzi F., Protti M.P., Sinigaglia F., Hammer J. Generation of tissue-specific and promiscuous HLA ligand databases using DNA microarrays and virtual HLA class II matrices. Nat. Biotechnol. 1999;17:555–561. doi: 10.1038/9858. [DOI] [PubMed] [Google Scholar]
- 26.Brusic V., Rudy G., Honeyman G., Hammer J., Harrison L. Prediction of MHC class II-binding peptides using an evolutionary algorithm and artificial neural network. Bioinformatics. 1998;14:121–130. doi: 10.1093/bioinformatics/14.2.121. [DOI] [PubMed] [Google Scholar]
- 27.De Groot A.S., Bosma A., Chinai N., Frost J., Jesdale B.M., Gonzalez M.A., Martin W., Saint-Aubin C. From genome to vaccine: in silico predictions, ex vivo verification. Vaccine. 2001;19:4385–4395. doi: 10.1016/s0264-410x(01)00145-1. [DOI] [PubMed] [Google Scholar]
- 28.Pruitt K.D., Tatusova T., Maglott D.R. NCBI Reference Sequence (RefSeq): a curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res. 2005;33:D501–D504. doi: 10.1093/nar/gki025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Boeckmann B., Bairoch A., Apweiler R., Blatter M.-C., Estreicher A., Gasteiger E., Martin M., Michoud K., O'Donovan C., Phan I., et al. The SWISS-PROT protein knowledgebase and its supplement TrEMBL in 2003. Nucleic Acids Res. 2003;31:365–370. doi: 10.1093/nar/gkg095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Altfeld M., Allen T.M., Yu X.G., Johnston M.N., Agrawal D., Korber B.T., Montefiori D.C., O'Connor D.H., Davis B.T., Lee P.K., et al. HIV-1 superinfection despite broad CD8+ T-cell responses containing replication of the primary virus. Nature. 2002;420:434–439. doi: 10.1038/nature01200. [DOI] [PubMed] [Google Scholar]
- 31.Allen T.M., Altfeld M., Yu X.G., O'Sullivan K.M., Lichterfeld M., Gall S.L., John M., Mothe B.R., Lee P.K., Kalife E.T., et al. Selection, transmission, and reversion of an antigen-processing cytotoxic T-lymphocyte escape mutation in human immunodeficiency virus type 1 infection. J. Virol. 2004;78:7069–7078. doi: 10.1128/JVI.78.13.7069-7078.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schuler M., Dönnes P., Nastke M.-D., Kohlbacher O., Rammensee H.-G., Stevanovic S. SNEP: SNP-derived Epitope Prediction program for minor H antigens. Immunogenetics. 2005;57:816–820. doi: 10.1007/s00251-005-0054-5. [DOI] [PubMed] [Google Scholar]