Abstract
HubMed is an alternative search interface to the PubMed database of biomedical literature, incorporating external web services and providing functions to improve the efficiency of literature search, browsing and retrieval. Users can create and visualize clusters of related articles, export citation data in multiple formats, receive daily updates of publications in their areas of interest, navigate links to full text and other related resources, retrieve data from formatted bibliography lists, navigate citation links and store annotated metadata for articles of interest. HubMed is freely available at http://www.hubmed.org/.
BACKGROUND
NCBI's PubMed (http://www.pubmed.gov/), a biomedical literature database incorporating MEDLINE, is the primary source of peer-reviewed biomedical information for scientific researchers, practising health professionals and the general public. Rapid response times from the search engine Entrez and integration with other NCBI-hosted databases such as GenBank allow PubMed to provide broad, up-to-date and curated search results. However, this breadth of coverage and functionality for a wide variety of users, ranging from those researching the results of clinical trials to those examining the composition of DNA sequences, means that Entrez/PubMed is unable to optimize its interface and functions for researchers that need to search and browse large volumes of literature covering their specific area of interest. The PubMed interface also lacks integration with web-based resources outside the NCBI.
Availability of the PubMed database via a web services API (http://www.ncbi.nlm.nih.gov/entrez/query/static/eutils_help.html), launched in 2002, opened up the possibility for external developers to take advantage of the NCBI's databases and processing power to provide alternative representations of the biomedical literature; e.g. analysing and extracting meaning from abstracts and MESH headings (1) or providing interfaces that add specialized functions (2).
FUNCTIONS PROVIDED BY HUBMED
HubMed (http://www.hubmed.org) is one such tool based around the Entrez Programming Utilities web service API. HubMed provides a dynamic and intuitive interface that transforms data from PubMed and integrates it with data from other sources, with the aim of improving the ability of researchers to find and manage biomedical literature related to their research.
For the last three years, HubMed has been providing daily updates of new arrivals to the MEDLINE database in a variety of XML (Extensible Markup Language) feed formats [currently Atom (http://atompub.org/), RSS 1.0 (RDF) and RSS 2.0 (http://blogs.law.harvard.edu/tech/rss)]. Subscribing to a feed of new matches for any search query is free and requires no registration, enabling tools such as Onfolio (http://www.onfolio.com/) and Kebberfegg (http://www.researchbuzz.org/tools/kebberfegg.pl) to dynamically generate feed subscriptions on demand, that can then be processed by desktop or web-based feed aggregators (see http://en.wikipedia.org/wiki/Aggregator for more details). Each item in a feed is linked via a unique identifier—the PubMed ID (PMID)—to HubMed's display of the most useful metadata available for that article, from where users can carry out a variety of functions, some of which are described below.
As most publications are not generally made available to researchers in a metadata-rich interchange format, the full text PDF of an article remains the most fundamental part of a researcher's digital library: an important link out of HubMed is therefore to the full online text of a paper. Users can proceed to the full text of an article using any of four overlapping options: through PubMed's ELink service (http://www.ncbi.nlm.nih.gov/entrez/query/static/elink_help.html) that leads to the document on the publisher's website; via Ex Libris' demonstration SFX server (http://www.exlibrisgroup.com/sfx/htm). that provides a range of alternate full text services (often based on either the PMID or Digital Object Identifier (DOI, http://www/doi.org/) of an article); through Google Scholar (http://scholar.google.com/), that carries out a full text search of selected web documents; or via activation of embedded COinS metadata (http://ocoins.info/) which allows anyone with a COinS-activating web browser extension (available from http://ocoins.info/#id3205609425) or proxy server to receive links to a full text resolver (based on the OpenURL linking standard, http://www.niso.org/standards/standard_detail.cfm?std_id=783) appropriate for their location or institutional affiliation.
While searching, browsing and reading articles, researchers are able to use HubMed to build a store of metadata for the papers that they find the most useful or interesting, as well as generating a taxonomy for these collections, by affixing tags (a synonym for keywords or labels) and annotations to each article. The Tag Storage service (http://www.hubmed.org/tags), which requires a free registration, facilitates the recall and browsing of articles collected by each user or user group. HubMed also works fluently with other academic- and science-targeted social bookmarking tools such as CiteULike (http://www.citeulike.org/) and Connotea (http://www.connotea.org/), both of which are able to automatically retrieve metadata for items stored using a PMID.
Once articles are stored inside HubMed's Tag Storage, users can arrange them into lists, view weighted visualizations of their tag usage frequency and export their stored data as RDF (http://www.w3.org/RDF/), for use with other tools. This RDF/XML export feature is also available from any HubMed search result page, providing a basis for the use of information harvesting and management tools, such as SIMILE's Piggy Bank (http://simile.mit.edu/piggy-bank/), an extension for the Firefox web browser that can be used to store, manipulate, browse and visualize data collected from any RDF data-exporting source. The possibilities enabled by this kind of semantic data store are numerous, such as inferring conflicts or agreements between networks of biomedical research publications (e.g. http://potlach.org/feast/2005/08/03/on-connecting-things/).
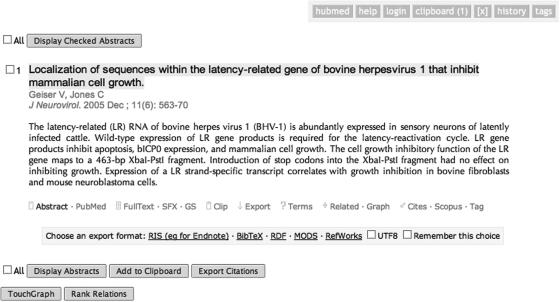
As illustrated in Figure 1, HubMed also provides direct export of article metadata in a range of other formats, including RIS (http://www.refman.com/support/risformat_intro.asp, for use with Endnote, RefDB and many other bibliographic tools), BibTeX (http://www.ecst.csuchico.edu/~jacobsd/bib/formats/bibtex.html, for use in TeX documents), MODS (http://www.loc.gov/standards/mods/, for use with XML document formats) and a direct link to send citation data to the online bibliographic library manager RefWorks (http://www.refworks.com/). HubMed maintains Unicode (UTF-8) characters throughout all its processes, so can provide the option to either include these accented characters in exported citation data or convert them to their Latin equivalents for use with older, Unicode-incompatible tools.
Figure 1.
A HubMed page displaying the abstract for a single article along with action links and options for a variety of export formats.
To aid researchers wishing to browse the bibliography lists of papers published online in PDF format, HubMed can extract bibliographic data from text copied and pasted from PDF documents. The Citation Finder, available at http://www.hubmed.org/citation.htm, extracts each reference, parses the citation string and converts it into a PubMed search; the results are then displayed in HubMed as standard search results, allowing users to continue to read and work with the referenced articles. This citation parsing algorithm is based on a modified version of the ParaTools Perl modules (http://paracite.eprints.org/developers/) produced by the Open Citation Project (http://opcit.eprints.org/).
To help users better understand jargon, acronyms and specialized scientific terms found within articles, HubMed's ‘Terms’ function, which accompanies each abstract, passes the abstract text through two web service filters in order to identify important keywords. The first, Whatizit (http://www.ebi.ac.uk/Rebholz-srv/whatizit/), is provided by the EBI and identifies Gene Ontology terms, along with protein and drug names in the text, adding links from each term to the Gene Ontology (3), UniProt (4) and MedlinePlus (http://medlineplus.gov/), respectively. The second filter compares all words to a database of Wikipedia page titles (available from http://download.wikipedia.org/) and adds links to the appropriate Wikipedia pages (http://www.wikipedia.org/) from words for which information is available. HubMed also aids search result browsing by extracting and displaying sentences from the abstract text in which the query terms occur. Additionally, searches are augmented both by the use of PubMed's ESpell web service (http://www.ncbi.nlm.nih.gov/entrez/query/static/espell_help.html), which provides alternative spelling suggestions for queries which return few or no results, and by a display of the MeSH categories (http://www.nlm.nih.gov/mesh/meshhome.html) matched by each query, which can be deselected or augmented as desired to refine the search query.
There are a number of tools in HubMed for exploring connections between related papers. Citation links can be explored directly for papers that are deposited in PubMed Central (data available from http://www.pubmedcentral.gov/utils/, including those from Open Access publisher BioMed Central), and there are also links to Elsevier's subscription service Scopus (http://www.scopus.com/), which allows in-depth exploration of citation and co-citation data. Articles related by co-occurrence of keywords can be explored directly as with normal search results using the relatedness score calculated by PubMed (described at http://www.ncbi.nlm.nih.gov/entrez/query/static/computation.html); these connections can be visualized as a dynamic force-directed graph using a TouchGraph Java applet (used with permission from http://www.touchgraph.com/). Articles can also be ranked by order of relatedness to multiple articles using HubMed's ‘Rank Relations’ feature, which allows an iterative refinement of clustered articles providing a more focused view of a topic than standard keyword searches. This is similar to a previously published process used for automatically updating bibliographies using ranking of related articles (5). In conjunction with browsing articles related by keywords and citation links, it would be useful to be able to browse the network of collaborations between authors of scientific papers (6), but this is currently precluded by a lack of unique author identifiers in the MEDLINE database, making it difficult to disambiguate multiple researchers who share the same name.
CONCLUSIONS
For future development, HubMed will continue to incorporate the functions of external web services as they become available (so far, all the mentioned web services have used simple Representational State Transfer (REST)-based interfaces), as well as augmenting built-in functions that improve search efficiency and user-friendliness. Personalization of searches and recommendations, based on patterns of user attention and implied interests, may also improve the accuracy of search results. The role of HubMed in providing building blocks for semantic life sciences data management will continue to adapt to new developments and the needs of researchers in this area.
Acknowledgments
The author would like to thank Gunther Eysenbach and Gurminder Bassi for critical reading of the manuscript and the reviewers for their helpful comments. This work is currently supported by the Journal of Medical Internet Research (http://www.jmir.org) and has previously received support from the University of Glasgow Division of Immunology, Infection and Inflammation. Funding to pay the Open Access publication charges for this article was provided by JMIR.
Conflict of interest statement. None declared.
REFERENCES
- 1.Perez-Iratxeta C., Pérez A.J., Bork P., Andrade M.A. Update on XplorMed: a web server for exploring scientific literature. Nucleic Acids Res. 2003;31:3866–68. doi: 10.1093/nar/gkg538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Doms A., Schroeder M. GoPubMed: exploring PubMed with the Gene Ontology. Nucleic Acids Res. 2005;33:783–86. doi: 10.1093/nar/gki470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ashburner M., Ball C.A., Blake J.A., Botstein D., Butler H., Cherry J.M., Davis A.P., Dolinski K., Dwight S.S., Eppig J.T., et al. Gene Ontology: tool for the unification of biology. The Gene Ontology Consortium. Nature Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Apweiler R., Bairoch A., Wu C.H., Barker W.C., Boeckmann B., Ferro S., Gasteiger E., Huang H., Lopez R., Magrane M., et al. UniProt: the Universal Protein knowledgebase. Nucleic Acids Res. 2004;32:115–19. doi: 10.1093/nar/gkh131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liu X., Altman R.B. Updating a bibliography using the related articles function within PubMed. Proc. AMIA Symp. 1998:750–54. [PMC free article] [PubMed] [Google Scholar]
- 6.Douglas S.M., Montelione G.T., Gerstein M. PubNet: a flexible system for visualizing literature derived networks. Genome Biol. 2005;6:R80. doi: 10.1186/gb-2005-6-9-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]