Abstract
Single nucleotide polymorphism (SNP) prioritization based on the phenotypic risk is essential for association studies. Assessment of the risk requires access to a variety of heterogeneous biological databases and analytical tools. FASTSNP (function analysis and selection tool for single nucleotide polymorphisms) is a web server that allows users to efficiently identify and prioritize high-risk SNPs according to their phenotypic risks and putative functional effects. A unique feature of FASTSNP is that the functional effect information used for SNP prioritization is always up-to-date, because FASTSNP extracts the information from 11 external web servers at query time using a team of web wrapper agents. Moreover, FASTSNP is extendable by simply deploying more Web wrapper agents. To validate the results of our prioritization, we analyzed 1569 SNPs from the SNP500Cancer database. The results show that SNPs with a high predicted risk exhibit low allele frequencies for the minor alleles, consistent with a well-known finding that a strong selective pressure exists for functional polymorphisms. We have been using FASTSNP for 2 years and FASTSNP enables us to discover a novel promoter polymorphism. FASTSNP is available at http://fastsnp.ibms.sinica.edu.tw.
INTRODUCTION
An important approach to disease gene mapping is investigating whether a single nucleotide polymorphism (SNP) is functionally involved in a disease. For complex diseases, the problem is complicated because, unlike Mendelian diseases, their genetic causes might involve many genes and hundreds of alleles. Although there are millions of SNPs deposited in public SNP databases, only a small proportion of them are functional polymorphisms that contribute to disease phenotypes. Thus, prioritizing SNPs based on their phenotypic risks is essential for association studies (1).
Assessment of the risk requires access to a variety of heterogeneous biological databases and analytical tools. FASTSNP (function analysis and selection tool for single nucleotide polymorphisms) is a web server that allows users to efficiently identify the SNPs most likely to have functional effects. It prioritizes SNPs according to 13 phenotypic risks and putative functional effects, such as changes to the transcriptional level, pre-mRNA splicing, protein structure and so on. A unique feature of FASTSNP is that the prediction of functional effects is always based on the most up-to-date information, which FASTSNP extracts from 11 external web servers at query time using a team of re-configurable web wrapper agents (2,3). These web wrapper agents automate web browsing and data extraction and can be easily configured and maintained with a tool that uses a machine learning algorithm. This allows users to configure/repair a web wrapper agent without programming. Another benefit of using web wrapper agents is that FASTSNP is extendable, so we can include new functions by simply deploying more web wrapper agents. In this manner, we have already built several new functionalities, such as the inclusion of information on haplotype blocks from HapMap (4).
SNP prioritization
Recent studies show that SNPs may have functional effects on the following.
transcriptional regulation, by affecting transcription factor binding sites in promoter or intronic enhancer regions (7,8); and
alternative splicing regulation, by disrupting exonic splicing enhancers (9) or silencers.
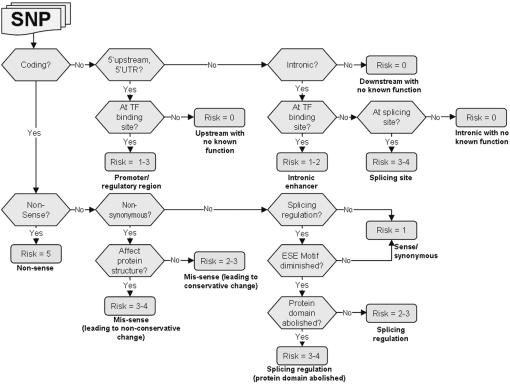
SNPs may also lead to premature termination of peptides (non-sense), which would disable the protein function. Each of these distinct functional effects may incur a risk that causes a disease. Therefore, to prioritize SNPs for the study of complex diseases, it is critical to identify the functional variants that are most likely to have functional effects leading to disease phenotypes before genotyping. Based on previous studies of the functional effects of polymorphisms, Tabor et al. (1) presented a prioritization strategy that associates the relative risk of a SNP with its location and the type of sequence variants. We extended their strategy with our recent findings and developed a decision tree to assess the risk of a SNP. The decision tree, shown in Figure 1, classifies a SNP into 1 of 13 types of the functional effects, each of which is assigned a risk ranking number between 0 and 5. A high risk rank implies a high-risk level. Table 1 gives the definitions of the function types, effects and their predicted risk ranking.
Figure 1.
Decision tree for prioritizing a SNP based on its functional effects. The diamonds represent decision points and the ovals represent terminal points with the risk and class assignments. Given an input SNP at a decision point, if the answer to the question in the diamond is ‘yes,’ then the vertical arrow should be followed. Otherwise, the horizontal arrow should be followed.
Table 1.
Definitions of the function types, their effects and predicted risks of SNPs
| Coding type | Function type | Possible effects | Risk (ranking) |
|---|---|---|---|
| Coding | Non-sense | Causes premature termination of an amino-acid sequence | Very high (5) |
| Splicing regulation (abolishing protein domain) | Breaks the exonic splicing enhancer/silencer binding site in a coding sequence, leading to abolished protein domain | Moderate to high (3∼4) | |
| Splicing regulation | Breaks the exonic splicing enhancer/silencer binding site in a coding sequence containing the same protein domains | Low to moderate (2∼3) | |
| Mis-sense (non-conservative change) | Alters an amino acid in a protein to one with different structure characteristics | Moderate to high (3∼4) | |
| Mis-sense (conservative change) | Alters an amino acid in a protein to one with similar structure characteristics | Low to moderate (2∼3) | |
| Sense/synonymous | Does not alter an amino acid in a sequence | Very low (1) | |
| Non-coding | Downstream with no known effect | No known effect | No known effect (0) |
| Upstream with no known effect | No known effect | No known effect (0) | |
| Splicing site | Breaks a consensus splicing site sequence | Moderate to high (3∼4) | |
| Promoter/regulatory region | Does not alter an amino acid, but can affect the level, location or timing of a gene expression | Very low to moderate (1∼3) | |
| Intronic enhancer | Alters a binding site of a transcription factor in an intronic region | Very low to low (1∼2) | |
| Untranslated | Changes an UTR in a sequence | No known effect to very low (0∼1) | |
| 3′utr post-transcriptional regulation | Breaks motifs likely to be involved in post-transcriptional regulation | Very low to moderate (1∼3) |
For a coding SNP, if it is non-synonymous and alters an amino acid in a protein resulting in a different protein structure (mis-sense, non-conservative change), or a non-sense change that results in a premature termination of the amino acid sequence (non-sense), then it will be assigned a high-risk or very high-risk ranking, because most known disease-causing SNPs are in these classes. A non-synonymous SNP that alters an amino acid in a protein to one with similar structural characteristics will be classified as ‘mis-sense, conservative change’ with a moderate risk ranking.
A coding SNP, be it synonymous or non-synonymous, may disturb the binding sites of an exonic splicing enhancer or silencer. In this case, the SNP may regulate alternative splicing and will be classified as ‘splicing regulation.’ If the alternative splicing further abolishes a domain of the translated protein due to exon skipping, then we classify such a SNP as ‘splicing regulation, abolishing protein domain’ and assign it a high-risk ranking. A ‘sense/synonymous’ SNP that does not affect any motif or protein structure will be assigned a low-risk ranking.
For a non-coding SNP, if its location is in the downstream region, it is unlikely that it has any functional effect. We classify this type of SNP as ‘downstream with no known function’ and assign no risk to it. However, if the transcription factor-binding site of the gene is changed by this polymorphism in the promoter region, then this SNP may affect the level, location, or timing of gene expression. In this case, it will be classified as ‘promoter/regulatory region’ with a low or moderate risk ranking; otherwise, it will be classified as ‘upstream with no known function.’ If it is in the intronic region, it may alter a binding site of the transcription factor and will be classified as ‘intronic enhancer’ with a low-risk ranking. A SNP at the ‘splicing site’ may break the consensus splicing site sequence and will thus be assigned a high-risk ranking.
A SNP at an untranslated region (UTR) of a sequence will be classified as ‘Untranslated’ with no risk. Recently, a new function type ‘3′-UTR post-transcriptional regulation’ was reported (10), but its actual phenotypic risk is still underdetermined. Since we have no substantial evidence about its risk, we will assign such a SNP a moderate-risk ranking. Consequently, our decision tree is complete in the sense that it considers all known functional roles of a SNP in a gene.
In addition to prioritizing on the basis of a SNPs' position and effect on a gene, Tabor et al. (1) further suggested filtering high-priority SNPs based on their allele frequencies, and using haplotype information to select a single SNP for genotyping if several SNPs are known to be in linkage disequilibrium. Though FASTSNP does not currently integrate this type of information in the prioritization method, it does provide their haplotype information in the output function report.
Web wrapper agents
The information required to answer the questions about the 13 decision points in the decision tree is available; however, it is spread over a variety of online sequence databases and analytical tools. Table 2 presents the databases and analytical tools used by FASTSNP for SNP prioritization and other functions. A traditional approach to integrating external sources is to download all the required data and programs and implement a huge data warehouse, but this often runs into maintenance difficulties because all the data must be kept up-to-date (11).
Table 2.
External web-based services accessed by FASTSNP
| Name/URL | Usage |
|---|---|
| NCBI dbSNP (www.ncbi.nlm.nih.gov/SNP) | Provides the location of a SNP in a gene and its alleles, allele frequency, and context sequence |
| Ensembl (www.ensembl.org) | Provides a cross-reference/alternative data source for dbSNP |
| TFSearch (www.cbrc.jp/research/db/TFSEARCH.html) | Predicts if a non-coding SNP alters the transcription factor-binding site of a gene |
| PolyPhen (www.bork.embl-heidelberg.de/PolyPhen) | Predicts if a non-synonymous SNP alters an amino acid in a protein resulting in structural changes (damaged or benign) in a protein |
| ESEfinder (rulai.cshl.edu/ESE) | Predicts if a synonymous SNP is located in a exonic splicing enhancer motif, which would diminish the motif with a different allele |
| RescueESE (genes.mit.edu/burgelab/rescue-ese) | Provides a cross-reference/alternative data source for ESEfinder |
| FAS-ESS (genes.mit.edu/fas-ess/) | Predicts exonic splicing silencer for each SNP allele |
| SwissProt (us.expasy.org/sprot) | Provides the information about protein domains to determine if a SNP causes an alternative splicing that leads to a protein domain being abolished |
| UCSC Golden Path (genome.ucsc.edu) | Provides information about the final draft assembly of the genome sequence (i.e. Golden Path) for quality control of candidate SNPs |
| NCBI Blast (www.ncbi.nlm.nih.gov/BLAST) | Sequence comparison and search tool for quality control of candidate SNPs |
| HapMap (www.hapmap.org) | Provides information about the haplotype and linkage disequilibrium around a SNP |
The first eight services provide databases and analytical tools to predict functional effects for SNP prioritization. UCSC Golden Path and NCBI Blast are used for quality control of candidate SNPs, while the haplotype database from HapMap is useful for further reducing the number of candidate genes for genotyping.
FASTSNP applies web wrapper agents (2,3) to resolve this problem. A web wrapper agent is a script that defines a user browsing session. When executed, the agent will visit the target website, fill in the query forms, extract the returned data and perform other web interactions to complete the user browsing session. Since FASTSNP uses the agent to access the external web servers at query time, the information used to prioritize SNPs is always the most up-to-date information available.
Web wrappers, however, are notorious for their fragility, because the format of web pages usually changes without notice. Previously, we developed a Java program, called Agent Toolbox, to cope with this issue (2). Agent Toolbox allows a user to produce an agent in a programming-by-example manner. More precisely, to produce an agent, the user simply browses the target website using the browser embedded in the user interface of Agent Toolbox to provide an example of a user session, and Agent Toolbox will generalize the example into a script that describes the user session.
Agent Toolbox can also generate data extraction rules to parse a given web page. Again, there is no need for users to program the data extraction rules. Instead, Agent Toolbox ‘learns’ these rules from users' labels on the web page with a machine learning algorithm (12,13). As a result, we can efficiently configure, repair and maintain the agents. Agent Toolbox also enables FASTSNP to be extended, so new functionalities can be included by deploying more web wrapper agents. For example, haplotype information is a new function that we included recently by producing an agent to access HapMap (4). Currently, a total of 11 agents are deployed by FASTSNP. A detailed explanation of how Agent Toolbox works is beyond the scope of this paper. For the most recent report, please refer to Ref. (2).
User interface
FASTSNP is available at http://fastsnp.ibms.sinica.edu.tw. Users can choose from three different methods to specify their SNPs as the input to query FASTSNP. The main query method is ‘Query by Candidate Gene.’ The user can choose to specify a gene symbol, SNP reference cluster ID (rsid), or a chromosome position as the query. Since the functional effects of an SNP may be different in different transcripts of the gene, FASTSNP will provide the transcripts of the queried gene for users to select before it outputs the set of SNPs on the queried gene. The user further refines the set of SNPs by specifying the region whether the SNPs are coding or non-coding and the allele frequency. After a final set of candidate SNPs is selected, FASTSNP will perform the SNP prioritization described earlier, return the prioritization results in a risk ranking report, and provide a function report for each candidate SNP. FASTSNP also provides ‘Query by SNP’ which allows the user to specify a single SNP rs ID or upload an Excel file containing their entire candidate SNPs for prioritization. Finally, FASTSNP also accepts novel SNP sequences as input. The user may paste a ‘novel SNP’ sequence in the text area and specify the position and the substitution via FASTSNP's web interface.
The function report on a SNP contains seven sections on the SNP's functional effects, namely (i) genomic information, presents the nearby sequence, the alleles and the allele frequency among different ethnic groups; (ii) functional effects summary, presents the risk assessment; (iii) transcription regulatory, shows the predicted transcription factor binding sites generated or disrupted by the different SNP alleles; (iv) alternative splicing regulatory, reports exonic splicing enhancer/silencer motifs changed by the SNP alleles leading to exon skipping or inclusion; (v) mRNA/protein domain effects, presents all spliced forms of mRNAs and protein variants extracted from GenBank (14). The protein domains that the SNP locates in are highlighted; (vi) protein structure effects, reports whether the SNP may cause a significant structural change in a protein; and (vii) SwissProt (15) feature table, provides information regarding other known mutations or variations of the translated protein of mRNAs related to the SNP. Some of these sections are specific to coding or non-coding SNPs and they will appear or not appear in the function report accordingly.
Implementation
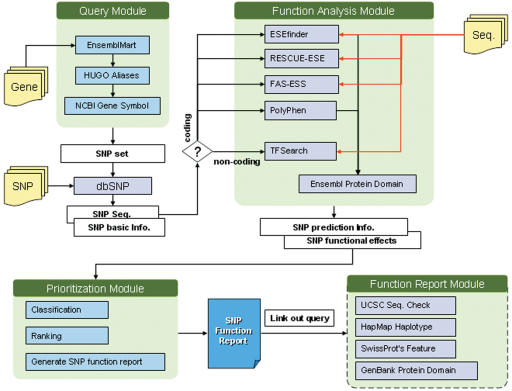
FASTSNP is written in Java and its web interface application is written in Java Server Pages (JSP). It runs on top of the Linux operating system and Tomcat web server and uses the MySQL database management system as its storage platform. Web wrapper agents are encapsulated as JavaBeans. Figure 2 shows the data flow diagram of the four major component modules in FASTSNP: (i) Query module, (ii) Function analysis module, (iii) Prioritization module and (iv) Function report module.
Figure 2.
FastSNP component modules and data flow. Each gray box represents an external web server accessed by a web wrapper agent, and each blue box represents a local Java program. Open boxes are data and the arrows represent the data flow.
The system will start at different modules when the user queries FASTSNP in different ways. When the user queries FASTSNP by gene, the query module will be used to check a cross-reference table and convert the query term to Ensembl Gene Name (16). The cross-reference table combines a set of widely used gene nomenclatures, including HUGO Gene Symbol, HUGO Gene Aliases (17), NCBI Reference Sequence (RefSeq) (18) and SwissProt Entry Name and users can query FASTSNP with these gene nomenclatures. With the converted gene name, we can extract a set of SNPs on the queried gene from the EnsemblMart database.
Given a set of SNPs, FASTSNP will dispatch the dbSNP agent to query dbSNP (19) for their genomic context sequences, their mRNA reference sequences (RefSeq), and their relative locations in gene sequences (e.g. 5′ upstream, 5′-UTR, non-synonymous, intronic and so on). Next, FASTSNP will prepare a set of sequences for each SNP as the input of the function analysis module. Each sequence is a 41 bp fragment trimmed from the genome sequence with the SNP allele exactly in the middle.
The function analysis module consists of three agent pipelines corresponding to decision paths in the decision tree shown in Figure 1. The first pipeline is for non-coding SNPs. The input sequence pair will be sent to TFSearch (20) to obtain the predicted transcription factor-binding sites. The second pipeline handles non-synonymous SNPs. In this pipeline, an agent will query PolyPhen (21) to obtain its prediction on whether the SNP will alter an amino acid in a protein and result in structural changes (damaged or benign) in the protein. The third agent pipeline obtains information to predict if the alternative splicing caused by a synonymous SNP may abolish a protein domain. FASTSNP will first invoke the agents for ESEfinder (22), RESCUE-ESE (23) and FAS-ESS (24) to predict if the SNP is located in an exonic splicing enhancer or silencer motif. It then invokes another agent to query Ensembl Protein Report (16) and obtain all known alternative spliced-form mRNAs for the gene where the SNP is located. With the spliced form mRNAs, the agent for SwissProt (15) will be able to extract the data about the translated proteins of the mRNAs to determine if the SNP leads to an alternative splicing that abolishes a protein domain. FASTSNP will perform the necessary post-processing for the data returned from the agent pipelines and submit the results to the prioritization module, which will then classify the SNP, assign it a risk ranking according to the decision tree shown in Figure 1, and compile the results into a function report.
In public domain databases, many SNPs are mapped to different positions in the human genome. Therefore in the function report, we provide buttons for users to invoke agents to check the sequence quality of SNPs and verify that a SNP uniquely maps to one position in the human genome. One of these agents maps the SNP on the UCSC Golden Path (25) sequence and integrates both NCBI and Ensembl annotations. FASTSNP also uses NCBI BLAST (26) to search the UCSC Golden Path to extend the SNP sequence to 500 base-pairs. We do this because a SNP's sequence in public domain databases is usually too short to allow the design of good quality primers.
FASTSNP also provides haplotype data, which is obtained from the HapMap (4). With haplotype information, we only need to select a minimum number of SNPs as markers (also known as tagSNPs) among the SNPs in linkage disequilibrium to conduct our association study. This reduces genotyping costs significantly.
Related works
Public domain SNP databases, such as dbSNP (19) and widely used SNP searching tools such as SNPper (27) contain well-organized catalogs of SNPs and provide entry portals to search for fundamental information about SNPs. However, searching for the information necessary to prioritize SNPs requires specialized Web servers, such as PupaSNP Finder (28), Wjst's system (29), PolyMAPr (30) and SNPselector (31). These servers usually integrate information from a variety of databases and analytical tools.
FASTSNP differs from the above systems in many respects. First, it uses a complete decision tree to assign risk rankings for SNP prioritization. It also considers some novel functional effects not considered by previous works, including the abolition of a protein domain due to exon skipping and the stability of a protein's structure. Another key feature of FASTSNP is that it applies web wrapper agents to query external sources at query time. As a result, it is highly extendable. Moreover, it always provides prioritization based on the most up-to-date information, thereby allowing users to update their queries. A recently published system called Galaxy (32) also aims at accomplishing this task. Their system provides up-to-date information by providing interactive communication to external websites to answer user queries.
Validation
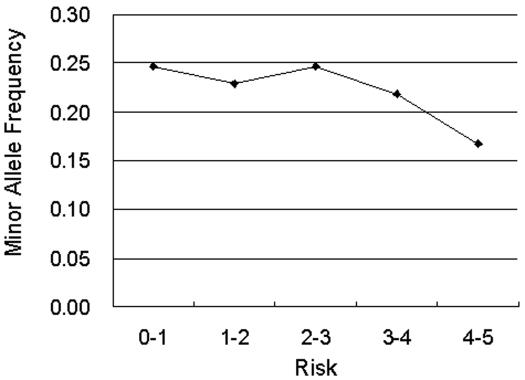
A well-known finding of functional polymorphisms is that they are usually under strong selective pressure and thus have low minor allele frequencies (5). To validate the results of our prioritization, we compared the output risk rankings of FASTSNP with the allele frequencies from SNP500Cancer (33) to see if SNPs with high predicted risks actually have low allele frequencies. The SNP500Cancer database was selected because it provides sufficient curated disease-causing SNPs with comparable allele-frequency results obtained by sequencing the same 102 subjects of different ethnic groups. FASTSNP successfully prioritized 1569 SNPs and the results, as given in Figure 3, show that high-risk SNPs tend to have low allele frequencies. SNPs with the highest risk have the least mean minor allele frequency (MAF) and MAF increases if the risk is not as high, consistent with Cargill et al.'s finding (5).
Figure 3.
Comparison of SNP's risk predicted by FASTSNP and their MAF. The results are obtained by analyzing 1569 SNPs from the SNP500Cancer database with FASTSNP.
RESULTS AND DISCUSSION
FASTSNP allows users to select functional polymorphisms for association studies in a convenient way. The National Genotyping Center (NGC) in Taiwan has been using this system to manage 5000 candidate SNPs and has already obtained their genotyping results using the MALDI-TOF high-throughput genotyping system. One of the most intensely researched topics at NGC is determining candidate genetic variants that contribute to adverse drug responses for different ethnic populations. Discovering major genetic factors that contribute to individual reactions to a certain medication may help to control and prevent side-effects or over dose during therapy, thereby avoiding serious consequences. With FASTSNP, the research team at NGC recently discovered a novel promoter polymorphism associated with the different therapeutic dosage of a drug (34). This polymorphism is predicted to change Ebox-binding site.
Acknowledgments
We wish to thank Po-He Tseng, Yuan-Chung Shen, Chih-Hung Kao, Siek Harianto and Sonny Wei for their help in the implementation of early version of FASTSNP, and Dr Shuen-Iu Hung, Dr Wu-Jer Yuarn and Dr Pei-Ing Hwang for their valuable suggestions. We also wish to thank the anonymous reviewers for their valuable comments. This project was supported in part by the National Research Program for Genomic Medicine (NRPGM), National Science Council, Taiwan, under Grant no. NSC94-3112-B-001-008-Y (National Genotyping Center) and Grant no. NSC94-3112-B-011-013-Y (Advanced Bioinformatics Core). Funding to pay the Open Access publication charges for this article was provided by the National Genotyping Center, Taiwan.
Conflict of interest statement. None declared.
REFERENCES
- 1.Tabor H.K., Risch N.J., Myers R.M. Opinion: candidate-gene approaches for studying complex genetic traits: practical considerations. Nature Rev. Genet. 2002;3:391–397. doi: 10.1038/nrg796. [DOI] [PubMed] [Google Scholar]
- 2.Hsu C.-N., Chang C.-H., Siek H., Lu J.-J., Chiou J.-J. Reconfigurable web wrapper agents for web information integration. IEEE Intell. Syst. 2003;18:34–40. [Google Scholar]
- 3.Hsu C.-N., Chang C.-H., Hsieh C.-H., Lu J.-J., Chang C.-C. Reconfigurable Web wrapper agents for biological information integration. J. Am. Soc. Info. Sci. Tech. 2005;56:505–517. [Google Scholar]
- 4.TheInternationalHapMapConsortium. The International HapMap Project. Nature. 2003;426:789–796. doi: 10.1038/nature02168. [DOI] [PubMed] [Google Scholar]
- 5.Cargill M., Altshuler D., Ireland J., Sklar P., Ardlie K., Patil N., Shaw N., Lane C.R., Lim E.P., Kalyanaraman N., et al. Characterization of single-nucleotide polymorphisms in coding regions of human genes. Nature Genet. 1999;22:231–238. doi: 10.1038/10290. [DOI] [PubMed] [Google Scholar]
- 6.Sunyaev S., Ramensky V., Bork P. Towards a structural basis of human non-synonymous single nucleotide polymorphisms. Trends Genet. 2000;16:198–200. doi: 10.1016/s0168-9525(00)01988-0. [DOI] [PubMed] [Google Scholar]
- 7.Prokunina L., Alarcn-Riquelme M.E. Regulatory SNPs in complex diseases: their identification and functional validation. Expert Rev. Mol. Med. 2004;2004:1–15. doi: 10.1017/S1462399404007690. [DOI] [PubMed] [Google Scholar]
- 8.Prokunina L., Castillejo-Lopez C., Oberg F., Gunnarsson I., Berg L., Magnusson V., Brookes A.J., Tentler D., Kristjansdottir H., Grondal G., et al. A regulatory polymorphism in PDCD1 is associated with susceptibility to systemic lupus erythematosus in humans. Nature Genet. 2002;32:666–669. doi: 10.1038/ng1020. [DOI] [PubMed] [Google Scholar]
- 9.Cartegni L., Krainer A.R. Disruption of an SF2/ASF-dependent exonic splicing enhancer in SMN2 causes spinal muscular atrophy in the absence of SMN1. Nature Genet. 2002;30:377–384. doi: 10.1038/ng854. [DOI] [PubMed] [Google Scholar]
- 10.Xie X., Lu J., Kulbokas E.J., Golub T.R., Mootha V., Lindblad-Toh K., Lander E.S., Kellis M. Systematic discovery of regulatory motifs in human promoters and 3′ UTRs by comparison of several mammals. Nature. 2005;434:338–345. doi: 10.1038/nature03441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stein L.D. Integrating biological databases. Nature Rev. Genet. 2003;4:337–345. doi: 10.1038/nrg1065. [DOI] [PubMed] [Google Scholar]
- 12.Hsu C.-N., Chang C.-H. Finite-state transducers for semi-structured text mining. In: Ronen Feldman., editor. Proceedings of IJCAI-99 Workshop on Text Mining: Foundations, Techniques and Applications; Menlo Park, CA, USA: IJCAI Co.; 1999. pp. 38–49. [Google Scholar]
- 13.Hsu C.-N., Dung M.-T. Generating finite-state transducers for semistructured data extraction from the web. Inform. Syst. 1998;23:521–538. [Google Scholar]
- 14.Benson D.A., Karsch-Mizrachi I., Lipman D.J., Ostell J., Wheeler D.L. GenBank: update. Nucleic Acids Res. 2004;32:D23–D26. doi: 10.1093/nar/gkh045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Boeckmann B., Bairoch A., Apweiler R., Blatter M.C., Estreicher A., Gasteiger E., Martin M.J., Michoud K., O'Donovan C., Phan I., et al. The Swiss-Prot protein knowledgebase and its supplement TrEMBL in 2003. Nucleic Acids Res. 2003;31:365–370. doi: 10.1093/nar/gkg095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Birney E., Andrews T.D., Bevan P., Caccamo M., Chen Y., Clarke L., Coates G., Cuff J., Curwen V., Cutts T., et al. An overview of Ensembl. Genome Res. 2004;14:925–928. doi: 10.1101/gr.1860604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Eyre T.A., Ducluzeau F., Sneddon T.P., Povey S., Bruford E.A., Lush M.J. The HUGO Gene Nomenclature Database, 2006 updates. Nucleic Acids Res. 2006;34:D319–D321. doi: 10.1093/nar/gkj147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pruitt K.D., Tatusova T., Maglott D.R. NCBI Reference Sequence (RefSeq): a curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res. 2005;33:D501–D504. doi: 10.1093/nar/gki025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sherry S.T., Ward M., Sirotkin K. dbSNP-database for single nucleotide polymorphisms and other classes of minor genetic variation. Genome Res. 1999;9:677–679. [PubMed] [Google Scholar]
- 20.Heinemeyer T., Wingender E., Reuter I., Hermjakob H., Kel A.E., Kel O.V., Ignatieva E.V., Ananko E.A., Podkolodnaya O.A., Kolpakov F.A., et al. Databases on transcriptional regulation: TRANSFAC, TRRD and COMPEL. Nucleic Acids Res. 1998;26:362–367. doi: 10.1093/nar/26.1.362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ramensky V., Bork P., Sunyaev S. Human non-synonymous SNPs: server and survey. Nucleic Acids Res. 2002;30:3894–3900. doi: 10.1093/nar/gkf493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cartegni L., Wang J., Zhu Z., Zhang M.Q., Krainer A.R. ESEfinder: a web resource to identify exonic splicing enhancers. Nucleic Acids Res. 2003;31:3568–3571. doi: 10.1093/nar/gkg616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fairbrother W.G., Yeo G.W., Yeh R., Goldstein P., Mawson M., Sharp P.A., Burge C.B. RESCUE-ESE identifies candidate exonic splicing enhancers in vertebrate exons. Nucleic Acids Res. 2004;32:W187–W190. doi: 10.1093/nar/gkh393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang Z., Rolish M.E., Yeo G., Tung V., Mawson M., Burge C.B. Systematic identification and analysis of exonic splicing silencers. Cell. 2004;119:831–845. doi: 10.1016/j.cell.2004.11.010. [DOI] [PubMed] [Google Scholar]
- 25.Karolchik D., Baertsch R., Diekhans M., Furey T.S., Hinrichs A., Lu Y.T., Roskin K.M., Schwartz M., Sugnet C.W., Thomas D.J., et al. The UCSC Genome Browser Database. Nucleic Acids Res. 2003;31:51–54. doi: 10.1093/nar/gkg129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McGinnis S., Madden T.L. BLAST: at the core of a powerful and diverse set of sequence analysis tools. Nucleic Acids Res. 2004;32:W20–W25. doi: 10.1093/nar/gkh435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Riva A., Kohane I.S. SNPper: retrieval and analysis of human SNPs. Bioinformatics. 2002;18:1681–1685. doi: 10.1093/bioinformatics/18.12.1681. [DOI] [PubMed] [Google Scholar]
- 28.Conde L., Vaquerizas J.M., Santoyo J., Al-Shahrour F., Ruiz-Llorente S., Robledo M., Dopazo J. PupaSNP Finder: a web tool for finding SNPs with putative effect at transcriptional level. Nucleic Acids Res. 2004;32:W242–W248. doi: 10.1093/nar/gkh438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wjst M. Target SNP selection in complex disease association studies. BMC Bioinformatics. 2004;5:92. doi: 10.1186/1471-2105-5-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Freimuth R.R., Stormo G.D., McLeod H.L. PolyMAPr: programs for polymorphism database mining, annotation, and functional analysis. Hum. Mutat. 2005;25:110–117. doi: 10.1002/humu.20123. [DOI] [PubMed] [Google Scholar]
- 31.Xu H., Gregory S.G., Hauser E.R., Stenger J.E., Pericak-Vance M.A., Vance J.M., Zuchner S., Hauser M.A. SNPselector: a web tool for selecting SNPs for genetic association studies. Bioinformatics. 2005;21:4181–4186. doi: 10.1093/bioinformatics/bti682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Giardine B., Riemer C., Hardison R.C., Burhans R., Elnitski L., Shah P., Zhang Y., Blankenberg D., Albert I., Taylor J., et al. Galaxy: a platform for interactive large-scale genome analysis. Genome Res. 2005;15:1451–1455. doi: 10.1101/gr.4086505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Packer B.R., Yeager M., Staats B., Welch R., Crenshaw A., Kiley M., Eckert A., Beerman M., Miller E., Bergen A., et al. SNP500Cancer: a public resource for sequence validation and assay development for genetic variation in candidate genes. Nucleic Acids Res. 2004;32:D528–D532. doi: 10.1093/nar/gkh005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yuan H.-Y., Chen J.-J., Lee M.-T., Wung J.-C., Chen Y.-F., Charng M.J., Lu M.-J., Hung C.-R., Wei C.-Y., Chen C.-H., et al. A novel functional VKORC1 promoter polymorphism is associated with inter-individual and inter-ethnic differences in warfarin sensitivity. Hum. Mol. Genet. 2005;14:1745–1751. doi: 10.1093/hmg/ddi180. [DOI] [PubMed] [Google Scholar]