Abstract
The herpes simplex viruses types 1 and 2 (HSV-1 and HSV-2) and varicella-zoster virus (VZV) can cause life-threatening infections of the central nervous system and lead to severe infections in immunocompromised subjects and newborns. In these cases, rapid diagnosis is crucial. We developed three different real-time PCR assays based on TaqMan chemistry for the LightCycler instrument to detect HSV-1, HSV-2, and VZV. When the TaqMan assays were compared to our in-house nested PCR assays, the test systems had equal sensitivities of ≤10 plasmid copies per assay. When clinical samples were investigated by TaqMan PCR to detect HSV-1, HSV-2, and VZV DNA, 95, 100, and 96% of the samples determined to be positive by nested PCR, respectively, were positive by the real-time PCR assays. The specificities of all PCR assays were almost 100%. Furthermore, the TaqMan PCR assays could be performed within 2.5 h, whereas nested PCR results were available after 9 h. In addition to offering more rapid results, the TaqMan PCR assays appear to be less expensive than nested PCR assays due to less hands-on time. In summary, TaqMan PCR is an excellent alternative to conventional nested PCR assays for the rapid detection of HSV-1, HSV-2, and VZV in clinical samples.
Herpes simplex viruses types 1 and 2 (HSV-1 and HSV-2) cause a variety of clinical symptoms in the central nervous system (CNS). In immunocompromised patients, the virus leads to severe clinical outcomes, including mucocutaneous disease and pneumonia. At delivery, HSV can be transmitted to the newborn and, following this exposure, can cause severe disseminated infections and death.
The varicella-zoster virus (VZV)—the causative agent of chicken pox and shingles—can also cause severe systemic infections of the CNS and the respiratory tract in immunocompetent individuals as well as in immunocompromised patients. The latter may also suffer from disseminated diseases of multiple organ systems.
Rapid laboratory diagnosis is urgently needed to distinguish HSV from VZV infections when the CNS is involved, especially in cases with clinically confusing dermal manifestations. Furthermore, rapid diagnosis of HSV and VZV infections is crucial in neonates to prevent a lethal outcome of disease (8, 9). Today, effective therapy of HSV and VZV infections is possible with antiviral drugs such as acyclovir. However, therapy must be initiated very early after the onset of disease to decrease lethality and to minimize the number of patients sustaining persistent neurological damage (15). Thus, the ability to rapidly diagnose HSV and VZV infections is a key feature in the virology laboratory. Molecular diagnostic assays using PCR are the standard for detecting herpesvirus infections of the CNS (1, 2, 10). Modifications of the basic PCR technique have been used to increase the sensitivity of detection of the viruses, e.g., by using nested PCR assays. Although very sensitive, this technique has the disadvantage of being highly susceptible to contamination, potentially leading to false-positive results (12).
Real-time PCR assays are based on the TaqMan chemistry or the HydProbe system (6, 7, 11, 16). Both systems combine a standard PCR with a fluorometer readout in a closed system. Real-time PCR also allows the detection of double-stranded DNA by SYBR Green technology. However, the use of individual TaqMan probes is superior to SYBR Green technology because TaqMan probe assays are more specific and lack false-positive results due to nonspecific amplification products, which are detected by SYBR Green technology.
Since real-time PCR allows a rapid detection of viral genomes in clinical specimens and is easy to use in a clinical laboratory, different real-time PCR assays for the detection of HSV and VZV have been developed recently (3-5, 9, 13).
The aim of this study was to develop three real-time PCR assays for the LightCycler instrument based on TaqMan chemistry for the detection of HSV-1, HSV-2, and VZV in clinical samples and to compare these assays to in-house nested PCR systems (1, 2).
MATERIALS AND METHODS
Clinical samples and viruses.
A total of 106 clinical samples, including 41 samples of cerebrospinal fluid (CSF), 28 swabs from patients with unknown skin diseases, 24 biopsy samples, 9 vitreous body samples, 3 blood samples, and 1 sample of bronchoalveolar lavage fluid, were obtained. All samples were collected mainly from immunocompromised subjects within the last 5 years and stored at −20°C. Among the total of 106 samples, 52 were obtained within the last 2 years. To study the specificities of the PCR assays, we investigated 15 clinical isolates of HSV-1, 12 isolates of HSV-2, and 11 isolates of VZV.
To analyze for clinical specificities of the PCR assays, we collected 25 samples from tumor patients or patients with chronic neurological disorders who were clinically not suspected to have HSV or VZV infections of the CNS.
Primers and probes.
Primers and probes were designed by using Primer Express 1.0 software (Applied Biosystems, Weiterstadt, Germany). Target genes were the glycoprotein D (gD) gene of HSV-1, the glycoprotein G (gG) gene of HSV-2, and the polymerase gene of VZV. The reporter (FAM) and the quencher (TAMRA) dyes were attached to the 5′ ends and the 3′ ends of the probes, respectively. Primer sequences, gene targets, and melting temperatures (in degrees Celsius) are listed in Table 1.
TABLE 1.
Primers and probes used for real-time PCR assays
| Virus | Primer or probea | Sequence (5′ to 3′) | Gene target | Tm (°C)b |
|---|---|---|---|---|
| HSV-1c | Forward primer (HSV1UP) | CGGCCGTGTGACACTATCG | gDc | 60 |
| Reverse primer (HSV1DP) | CTCGTAAAATGGCCCCTCC | gD | 60 | |
| Probe (HSV1P) | CCATACCGACCACACCGACGAACC | gD | 70 | |
| HSV-2d | Forward primer (HSV2UP) | CGCTCTCGTAAATGCTTCCCT | gGd | 60 |
| Reverse primer (HSV2DP) | TCTACCCACAACAGACCCACG | gG | 60 | |
| Probe (HSV2P) | CGCGGAGACATTCGAGTACCAGATCG | gG | 70 | |
| VZVe | Forward primer (VZV UP) | CGGCATGGCCCGTCTAT | DNA polymerasee | 60 |
| Reverse primer (VZV DP) | TCGCGTGCTGCGGC | DNA polymerase | 60 | |
| Probe (VZV P) | ATTCAGCAATGGAAACACACGACGCC | DNA polymerase | 70 |
All probes were labeled with FAM at the 3′ end and with TAMRA at the 5′ end.
The melting temperature (Tm) was calculated by using the Primer Express software based on the Rychlik algorithm.
Gene bank accession no. AF141854-58, AJ303204, and Z86099.
Gene bank accession no. AB059828-31 and X04370.
DNA standards.
For each PCR assay, we cloned the amplicon target-DNA fragment into the vector pCRII by using the TA cloning kit (Invitrogen, Breda, The Netherlands). The following plasmids were generated: pCRIIHSV1, containing 70 bp of the gD gene; pCRHSV2, containing 111 bp of the gG gene; and pCRIIVZV, containing 63 bp of the VZV DNA polymerase gene. Standards of 104 to 100 genome copies per 5 μl of water were prepared from linearized plasmids for each assay. For the nested PCR assay controls, we cloned the first-round amplification products into pCRII by using the TA cloning kit. These plasmids were diluted, and they served as standards to determine the sensitivities of the assays.
DNA extraction.
DNA was extracted from 140 μl of clinical specimens by using the QIAmp DNA extraction kit (Qiagen, Hilden, Germany) according to the manufacturer's instructions. DNA was recovered in 50 μl of water, and 5 μl was immediately analyzed by real-time PCR, while the remaining 45 μl was stored at −20°C. When DNA was prepared from blister fluid or swabs, a single-sample preparation procedure was followed to avoid cross contamination of samples prepared in parallel.
Nested PCR.
Three nested PCR assays to detect HSV-1, HSV-2, and VZV were developed. To detect HSV-1, we used the nested PCR protocol by Aurelius et al. (2) with the primers BJ HSV1.1 and BJ HSV1.2 for the first-round amplification and the primers BJ HSV1.3 and BJ HSV1.4 for the second-round amplification. The primers were located in the gD gene (2). To detect HSV-2 infections, we applied a second nested PCR protocol (1) with the primers BJ HSV2.1 and BJ HSV2.2 for the first-round amplification and the primers BJ HSV2.3 and BJ HSV2.4 for the second round; both sets are also located in the gG gene (1).
To detect VZV, we used an amplicon targeting the DNA polymerase. The first-round primers (VZV1outhu, 5′ CCC GCG GTG GAG ACG ACT T 3′, and VZV2outhu, 5′ ATT GCG GGG TTG GGT GAG C 3′) amplified a fragment of 694 bp. The second-round primers (VZV3 inhu, 5′ GCG GTG GAG ACG ACT TCA ATA GCA 3′, and VZV4 inhu, 5′ TGG AGG AAG AGA CGT GGA GAC TGG 3′) amplified a 257-bp fragment. To detect HSV-1, HSV-2, and VZV, we used a single protocol. Amplifications were performed in a total volume of 50 μl by using 10 μl of prepared DNA for the first round and 1 μl of the first-round sample for nested amplification. Amplification was carried out in AmpliTaq Gold PCR Mix supplemented with 2.5 mM MgCl2 by using 10 pmol of primers in the first round and 50 pmol of primers in the second round. The cycle conditions were identical for both rounds. The AmpliTaq Gold was activated at 94°C for 10 s. Then, 30 cycles were performed by using the following profile: 10 cycles of 94°C for 30 s, 65°C for 30 s, and 72°C for 30 s; 10 cycles of 94°C for 50 s, 65°C for 30 s, and 72°C for 30 s; and 10 cycles of 94°C for 80 s, 65°C for 30 s, and 72°C for 30 s. The PCR products were visualized by using a 2% agarose gel stained with ethidium bromide. In every PCR assay, five no-template controls were included in each nested PCR run.
Real-time PCR.
Three different real-time PCR assays based on TaqMan chemistry to detect HSV-1, HSV-2, and VZV were developed. All real-time PCR assays were performed in the same format and were run on the LightCycler (Roche, Mannheim, Germany). Five microliters of the plasmid standard or of DNA prepared from clinical specimens was added to 15 μl of LightCycler FastStart-DNA-Master-Hybridization-Probes reaction mix (Roche) supplemented with 3.2 mM MgCl2, 300 nM (each) primers, 200 nM corresponding probe, and 1 U of uracil-DNA glycosylase (Roche). Primers and probes were applied as listed in Table 1. The cycle conditions were identical for all three PCR assays. To degrade potential contaminating products, a uracil-DNA glycosylase step (20°C for 600 s, 95°C for 300 s) was performed, followed by 45 amplification cycles (95°C for 15 s, 60°C for 30 s; heating rate, 20°C/s).
Prevention of PCR contamination.
Nested PCR systems are known to be error prone due to random contamination (12). The risk of contamination was reduced by introducing a one-way working scheme in which the preparation of reagents, sample preparation, and first-round and second-round amplifications were carried out in different rooms and in different buildings with separate air-conditioning systems. Furthermore, special clothing and nurses' caps were worn in the different working areas. To remove potential DNA contamination from the racks used in the reagent and sample preparation area, we immersed the racks for 1 h in 1 N HCl and drained them. When the real-time PCR technology was applied, we kept to the three-room system (reagent preparation, sample preparation, and amplification) as described above and included a contamination protection step by using the uracil-DNA glycosylase carryover prevention system.
RESULTS
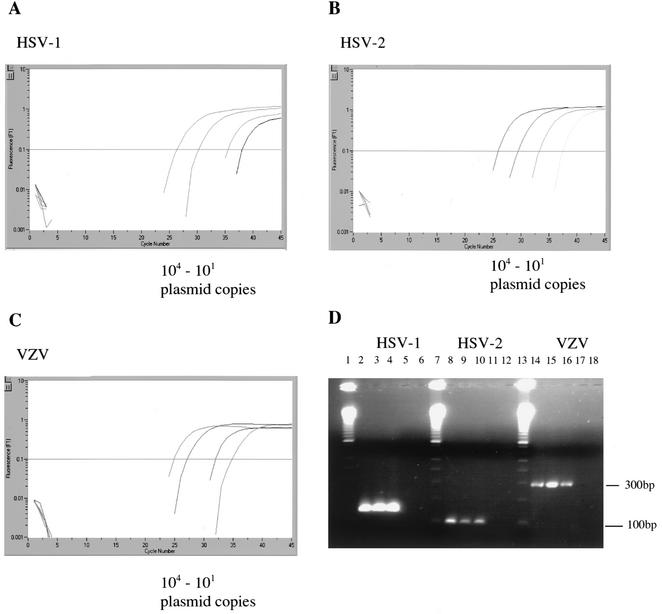
When 10-fold dilutions of the plasmids were analyzed on the LightCycler instrument by using TaqMan real-time PCR assays, the detection limit was 10 copies per reaction for HSV-1, HSV-2, and VZV standard DNA. This was confirmed by the fact that, when fewer than 10 plasmid copies were analyzed by nested PCR or real-time PCR, both systems detected those samples infrequently (data not shown). As outlined in Fig. 1, there was no difference in the sensitivities when real-time PCR assays were compared to our traditional nested PCR systems. We analyzed 15 isolates of HSV-1, 12 isolates of HSV-2, and 11 isolates of VZV from our strain collection by both systems and all three assays (HSV-1, HSV-2, and VZV). All isolates were positive by the corresponding PCR assay, indicating that these assays were highly specific across the range of our strain collection. Specificities of the PCR assays and systems appeared to be 100%, since neither the real-time PCR assays nor the conventional nested PCR assays showed any cross-reaction when noncorresponding herpesvirus DNA and primer sets were analyzed. To evaluate the clinical specificities of the two PCR systems, we analyzed 25 CSF samples from patients clinically not suspected of having HSV or VZV CNS infections. None of these samples were positive with the real-time PCR assays or the nested PCR assays. To compare the real-time PCR assay with conventional nested PCR, we analyzed clinical samples that tested positive by nested PCR for either HSV-1, HSV-2, or VZV. Samples determined to be positive by nested PCR were collected over a time period of 5 years, and sample DNA was stored at −20°C. From 46 HSV-1 samples pretested for HSV-1 and determined to be positive by nested PCR, 40 (87%) turned out to be positive by the real-time PCR assay. These samples included 21 CSF samples from patients with encephalitis, 5 swabs from patients with unknown skin diseases, 15 biopsy samples (including bone marrow, lung, and biopsy specimens from the gastrointestinal tract), 1 sample of bronchoalveolar lavage fluid, 1 vitreous body sample, and 3 blood samples. Two blood samples were obtained from AIDS patients, and one was obtained from an HSV-1-infected newborn. When the six samples which were negative for HSV-1 DNA by the real-time PCR assay were retested by nested PCR, four turned out to be negative. Thus, HSV-1 real-time PCR detected 95% of the samples that tested positive by nested PCR.
FIG. 1.
Sensitivities of the PCR assays for detecting HSV-1, HSV-2, and VZV. (A to C) Results of real-time PCR. Plasmids were serially diluted from 104 to 101 copies per assay. In the lower left parts of the graphs, results for the corresponding no-template controls are shown. Vertical (y) axis, fluorescence intensity; horizontal (x) axis, PCR cycle numbers. (D) Results of nested PCR to detect HSV-1, HSV-2, and VZV. Lane 1, molecular weight marker (MWM); lanes 2 to 4, 103 to 101 copies of HSV-1 plasmid controls; lanes 5 and 6, no-template control; lane 7, MWM; lanes 8 to 10, 103 to 101 copies of HSV-2 plasmid controls; lanes 11 and 12, no-template control; lane 13, MWM; lanes 14 to 16, 103 to 101 copies of VZV plasmid controls; lanes 17 and 18, no-template control.
Only four clinical samples tested positive for HSV-2 DNA by nested PCR. These included one CSF sample from a case of encephalitis, two vitreous body samples from cases with clinical diagnoses of uveitis, and one genital swab. All samples tested were positive by the real-time PCR assay.
Fifty-six clinical samples that previously tested positive for VZV DNA by nested PCR included the following: 19 CSF samples from patients with myeloencephalitis, 6 vitreous body samples obtained from patients with acute necrosis of the retina, 22 swabs from patients with unknown skin lesions, and 9 tissue biopsy specimens. In 54 (96%) of 56 samples that tested positive by nested PCR, VZV DNA was detected by real-time PCR. Retesting of the negative samples by nested PCR gave the same results.
DISCUSSION
We evaluated three different real-time PCR assays to detect HSV-1, HSV-2, and VZV and compared the assays with conventional nested PCR assays. The nested PCR assays used in this study to detect HSV-1 and HSV-2 were performed according to a protocol published by Aurelius et al. (1, 2), with modifications, yielding sensitivities at least similar to those published. Although 106 clinical samples were investigated, only four samples turned out to be positive for HSV-2. Thus, the low number of HSV-2-positive clinical samples does not really permit a valid assessment of the HSV-2 assay.
There was no difference in the detection limits of the real-time PCR assays and the corresponding nested PCR test system when plasmid DNA was used. Both test systems were highly sensitive and highly specific, with the sensitivities and specificities of our real-time PCR assays concordant with those reported in other studies (3-5, 9, 13). The fact that real-time PCR was not able to detect viral DNA in all clinical samples when compared to nested PCR might be due to the fact that some samples may have degraded over the 5-year experimental period. Thus, sample degradation may easily explain why not all samples were positive in the real-time assays when tested retrospectively after years of storage. Since sample preparation was uniform for both types of assays, inhibitors present in the DNA preparations could not explain the different results obtained when both PCR systems were compared.
Nested PCR systems are known to be error prone due to random contamination (12). Contamination could not be excluded because, after DNA preparation, multiple pipetting steps are required between first-round and second-round amplifications. To reduce the risk of contamination, a one-way working scheme was used as described in Material and Methods. For the real-time PCR assays, we included a contamination protection step by using the uracil-DNA glycosylase carryover prevention system. Furthermore, the tubes were not opened at any time during the amplification procedure. Thus, contamination potentially appears to be a very rare event in the real-time PCR assays, if it occurs at all.
One further advantage of the real-time PCR assays described in this study is that they all were performed with the same amplification protocol. Thus, within one LightCycler run, a sample can be tested for HSV-1, HSV-2, and VZV and all positive and negative controls for each virus. This has two major benefits: (i) in emergency cases in which rapid diagnosis is important, the laboratory can run the assays simultaneously with only one LightCycler instrument, and (ii) in the routine setting, samples may be analyzed for HSV or VZV independently. Although the costs for consumables are higher for real-time assays than for nested PCR assays, the total costs for real-time PCR assays are lower due to less hands-on time and the optimal use of reagents and instruments, and with less sample manipulation there should be fewer false-positive results.
In many clinical cases, a very rapid diagnosis is crucial, and as shown above, real-time PCR has the advantage of being much faster than nested PCR systems. In our hands, viral DNA was detected by real-time PCR within 2.5 h, which included DNA preparation from clinical specimens. The nested PCR assays took about 9 h, making the real-time PCR assays 3.6 times faster than the nested assays. In summary, the TaqMan assays performed on the LightCycler are suitable tools for the rapid diagnosis of HSV-1, HSV-2, and VZV infections in the clinical virology laboratory. Due to the ease of use, the significant decrease in testing and handling time, and the high speed of performance, real-time PCR assays can be used in the laboratory, even in emergency cases in which rapid diagnosis is essential.
Acknowledgments
We thank Heike Schley for excellent technical assistance.
This work was supported in part by the Bundesministerium für Bildung und Forschung (BMBF) grant no. 01KI9951.
REFERENCES
- 1.Aurelius, E., B. Johansson, B. Skoldenberg, and M. Forsgren. 1993. Encephalitis in immunocompetent patients due to herpes simplex virus type 1 or 2 as determined by type-specific polymerase chain reaction and antibody assays of cerebrospinal fluid. J. Med. Virol. 39:179-186. [DOI] [PubMed] [Google Scholar]
- 2.Aurelius, E., B. Johansson, B. Skoldenberg, A. Staland, and M. Forsgren. 1991. Rapid diagnosis of herpes simplex encephalitis by nested polymerase chain reaction assay of cerebrospinal fluid. Lancet 337:189-192. [DOI] [PubMed] [Google Scholar]
- 3.Espy, M. J., R. Teo, T. K. Ross, K. A. Svien, A. D. Wold, J. R. Uhl, and T. F. Smith. 2000. Diagnosis of varicella-zoster virus infections in the clinical laboratory by LightCycler PCR. J. Clin. Microbiol. 38:3187-3189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Espy, M. J., J. R. Uhl, P. S. Mitchell, J. N. Thorvilson, K. A. Svien, A. D. Wold, and T. F. Smith. 2000. Diagnosis of herpes simplex virus infections in the clinical laboratory by LightCycler PCR. J. Clin. Microbiol. 38:795-799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Furuta, Y., F. Ohtani, H. Sawa, S. Fukuda, and Y. Inuyama. 2001. Quantitation of varicella-zoster virus DNA in patients with Ramsay Hunt syndrome and zoster sine herpete. J. Clin. Microbiol. 39:2856-2859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Heid, C. A., J. Stevens, K. J. Livak, and P. M. Williams. 1996. Real time quantitative PCR. Genome Res. 6:986-994. [DOI] [PubMed] [Google Scholar]
- 7.Holland, P. M., R. D. Abramson, R. Watson, and D. H. Gelfand. 1991. Detection of specific polymerase chain reaction product by utilizing the 5′—3′ exonuclease activity of Thermus aquaticus DNA polymerase. Proc. Natl. Acad. Sci. USA 88:7276-7280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hufert, F. T., T. Diebold, B. Ermisch, D. von Laer, U. Meyer-König, and D. Neumann-Haefelin. 1995. Liver failure due to disseminated HSV-1 infection in a newborn twin. Scand. J. Infect. Dis. 27:627-629. [DOI] [PubMed] [Google Scholar]
- 9.Kessler, H. H., G. Muhlbauer, B. Rinner, E. Stelzl, A. Berger, H. W. Dorr, B. Santner, E. Marth, and H. Rabenau. 2000. Detection of herpes simplex virus DNA by real-time PCR. J. Clin. Microbiol. 38:2638-2642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Koskiniemi, M., H. Piiparinen, L. Mannonen, T. Rantalaiho, A. Vaheri, and the Study Group. 1996. Herpes encephalitis is a disease of middle aged and elderly people: polymerase chain reaction for detection of herpes simplex virus in the CSF of 516 patients with encephalitis. J. Neurol. Neurosurg. Psychiatry 60:174-178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee, L. G., C. R. Connell, and W. Bloch. 1993. Allelic discrimination by nick-translation PCR with fluorogenic probes. Nucleic Acids Res. 21:3761-3766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Porter-Jordan, K., E. I. Rosenberg, J. F. Keiser, J. D. Gross, A. M. Ross, and S. Nasim. 1990. Nested polymerase chain reaction assay for the detection of cytomegalovirus overcomes false positives caused by contamination with fragmented DNA. J. Med. Virol. 30:85-91. [DOI] [PubMed] [Google Scholar]
- 13.Ryncarz, A. J., J. Goddard, A. Wald, M. L. Huang, B. Roizman, and L. Corey. 1999. Development of a high-throughput quantitative assay for detecting herpes simplex virus DNA in clinical samples. J. Clin. Microbiol. 37:1941-1947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Whitley, R. J. 2001. Herpes simplex viruses, p. 2461-2509. In D. N. Knipe and P. M. Howley (ed.), Fields Virology, 4th ed., vol. 2. Lippincott Williams & Wilkins, Baltimore, Md.
- 15.Whitley, R. J., D. W. Kimberlin, and B. Roizman. 1998. Herpes simplex viruses. Clin. Infect. Dis. 26:541-555. [DOI] [PubMed] [Google Scholar]
- 16.Wittwer, C. T., M. G. Herrmann, A. A. Moss, and R. P. Rasmussen. 1997. Continuous fluorescence monitoring of rapid cycle DNA amplification. BioTechniques 22:130-131, 134-138. [DOI] [PubMed]