Abstract
Protein–DNA interactions play a central role in regulatory processes at the genetic level. DNA-binding proteins recognize their targets by direct base–amino acid interactions and indirect conformational energy contribution from DNA deformations and elasticity. Knowledge-based approach based on the statistical analysis of protein–DNA complex structures has been successfully used to calculate interaction energies and specificities of direct and indirect readouts in protein–DNA recognition. Here, we have implemented the method as a webserver, which calculates direct and indirect readout energies and Z-scores, as a measure of specificity, using atomic coordinates of protein–DNA complexes. This server is freely available at http://gibk26.bse.kyutech.ac.jp/jouhou/readout/. The only input to this webserver is the Protein Data Bank (PDB) style coordinate data of atoms or the PDB code itself. The server returns total energy Z-scores, which estimate the degree of sequence specificity of the protein–DNA complex. This webserver is expected to be useful for estimating interaction energy and DNA conformation energy, and relative contributions to the specificity from direct and indirect readout. It may also be useful for checking the quality of protein–DNA complex structures, and for engineering proteins and target DNAs.
INTRODUCTION
Gene regulatory proteins such as transcription factors recognize DNA sequences through direct interactions between amino acids and base pairs (direct readout) and/or through specific conformations and elastic properties of DNA (indirect readout) (1–7). In order to understand the mechanism of protein–DNA recognition, we need to quantify the interaction and conformation energies and relative contributions of direct and indirect readouts to the specificity. In general, there are two kinds of approaches to this problem. One is the knowledge-based approach using known protein–DNA complex structures to calculate statistical potentials and specificities (2,5,8,9). The other is the ab initio approach based on computer simulations to calculate the energies and specificities (10–17). Both approaches have advantages and disadvantages, and would complement each other. In either approach, we need to perform two different types of energy calculations. One of them is the calculation of elastic deformations of DNA, in which a DNA molecule is treated as an elastic object, with several degrees of freedom in its conformations for the indirect readout. The other is that of base–amino acid interactions between protein and DNA for the direct readout.
In the knowledge-based approach, we have calculated statistical potentials for base–amino acid interactions and sequence-dependent conformation of DNA, and estimated normalized energy Z-scores as a measure of specificity for a given protein–DNA complex by using a sequence-structure threading method (8,9). The role of solvent appears implicitly in the calculations, as the knowledge-based potentials are based on protein–DNA complexes, including solvent and other thermodynamic effects. Using this method, we have been able to successfully explain the role of direct and indirect readout contributions in protein–DNA recognition (5), and to predict target DNA sequences (8,18). Calculations of these quantities for a given protein–DNA complex are almost as fast as sequence-based prediction methods. Here, we present a webserver, which can be used to obtain the energy Z-scores based on our force fields. This webserver takes the coordinate data of a protein–DNA complex and calculates its conformational parameters and base-amino acid contacts. It may be noted that only the DNA conformational parameters are taken into account, because we consider here the specificity to a given protein (a fixed amino acid sequence) with changing DNA sequences, and because the conformational change of a protein, if any, will be within the current direct potential resolution of a 3 Å cubic grid. However, the magnitude of the protein conformational change corresponding to distinct DNA sequences could be larger than that and it remains to be analyzed. It is expected that users studying the protein–DNA recognition will find this server a crucial tool to analyze the interaction energy, the DNA conformation energy, and the relative contributions to the specificity from direct and indirect readouts, to check the quality of protein–DNA complex structures, and to engineer proteins and target DNAs.
MATERIALS AND METHODS
Direct readout energy
Protein residues and DNA bases show a variety of interactions. Some interactions such as Asn-A and Lys-G are frequently observed in the complex structures. The spatial distributions of side chains around base pairs indicate the possibility that the distribution may be converted to energy potential, in a manner similar to the contact potential between amino acids in protein structures, and it can be used for the target prediction (8). In order to derive the statistical potential of interactions between bases and amino acids, we defined a coordinate system by taking an origin at the N9 atom for A and G, and at the N1 atom for T and C. We considered the amino acids within a given box, and the box was divided into grids. Then we transformed the distributions of the Cα atom into statistical potentials defined by the following equations:
where mab is the observed number of pairs a and b, w is the weight given to each observation - taken as 20 for the current calculations, f(s) is the relative frequency of occurrence of any amino acid at the grid point s, and gab(s) is the equivalent relative frequency of occurrence of amino acid a against base b. R and T are the gas constant and the absolute temperature, respectively. Here, we used a box of ∣x∣ = ∣y∣ = 13.5 Å and ∣z∣ = 6 Å, and a grid interval of 3 Å, which was optimized by examining various intervals. Energy scores were normalized against random DNA sequences to obtain the Z-scores as described in Energy Z-scores section.
Indirect readout energy
Conformational parameters of DNA
There are many ways in which DNA conformation may be characterized. Our force fields are based on six types of conformational parameters, namely shift, slide, rise, tilt, roll and twist (19–21). The values of these parameters are extracted from the output of the 3DNA program provided by Olson group (19).
Development of force field
Conformational energy of a particular base pair depends on the deformation at that base pair position for each type of conformation. As an example, Figure 1 schematically shows the deformation in a base pair with respect to the mean or expected angle of tilt. Elasticity of the given base pair conformation for any tilt can be extracted from the distribution of tilt angles in the whole database. In this webserver, the force field used is based on our previously published work (5). In terms of energy, E, the Z-score is calculated under the hypothesis that the conformational energy is a harmonic function of the conformational coordinates following the expression (2):
where kB is the Boltzmann constant and T the absolute temperature (for the purpose of Z-scores calculation, kBT can be taken equal to one, as the energy comparison is made in arbitrary units), is the conformational parameter of the coordinate i of the base step s. The elements of each force field matrix Fs are calculated through matrix inversion of the covariance matrix of each base step s of a reference dataset of non-redundant protein–DNA complexes (9). Finally the conformation energy of a structure is given as follows:
where ΔΘs is the six dimensional conformational fluctuation of the base step s and Fs is the force field matrix associated with base step s.
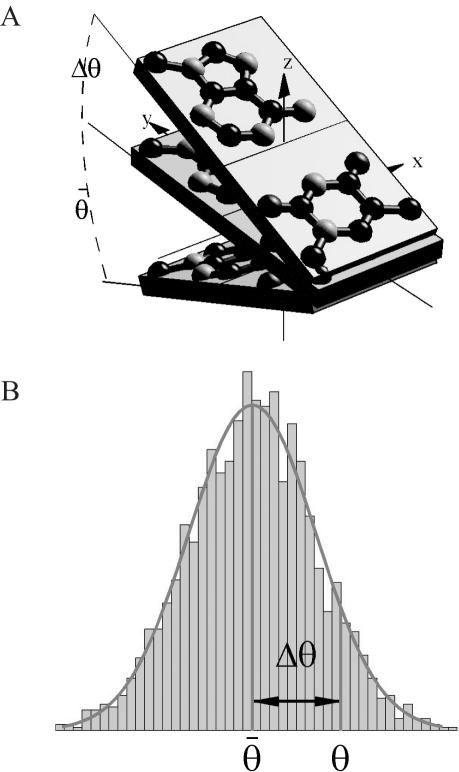
Figure 1.
(a) An example of the conformational parameter (tilt). Two successive bases in the DNA helix are tilted to each other by an angle. θ represents the mean deformation in the database (used for developing the force field) and Δθ shows the deformation in a base pair for an example target. Energy contribution from this base pair deformation depends on the overall distribution of θ in the database. (b) A typical distribution of elastic deformation values in the DNA.
Energy Z-scores
The energy Z-score for a target sequence and structure determines the specificity of that sequence towards the observed conformation or structure. The Z-score or the specificity of a DNA sequence is calculated in terms of the energy of the target sequence observed in the given protein–DNA complex against a set of random sequences. Detailed procedures of computing these scores are defined in our earlier work (8). Here it may suffice to mention that a number of random DNA sequences having the same length as the target sequence are generated. These sequences are assumed to form exactly the same structure as the target complex and their energies of such hypothetical complexes are calculated. The Z-score is then defined as follows:
where X is the energy of a particular sequence, μ is the mean energy of 50 000 (or any other selected number of) random DNA sequences, and σ is the standard deviation (SD) of these random sequences. A more negative Z-score implies that a target sequence fits better to the given structure. The ReadOut webserver will output the energy Z-scores for indirect and direct readout energies.
Selection of base pairs
Base pair conformational parameters show anomalous behavior at the terminal and kinked positions. The program 3DNA (19) is used for filtering out those bases that do not form base pairs. The server automatically obtains this information about base pairing and determines which base numbers are to be included in the calculations. A separate table parsed from 3DNA output is provided as the query results, which supplies information about base pairs and the region of the DNA involved.
Query submission and output
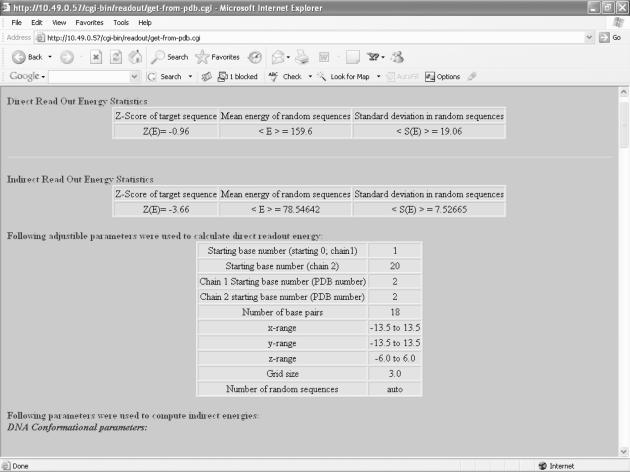
Protein Data Bank (PDB) style coordinate file is the only input to the server. Alternatively, a four-letter PDB code may be entered if the corresponding structural data are available in PDB (22). An example output results page of the server is shown in Figure 2. These results consist of the following:
Indirect and direct readout Z-scores, mean energy and SDs.
Conformational parameters: In the end, we provide the data of the conformational parameters in each base steps used to obtain the reported Z-scores and additional information. These values are calculated using the 3DNA program (19).
Figure 2.
Snapshot of typical output of the ReadOut webserver.
PDB data and biological units
Information about the atomic coordinates in PDB faces difficulties as the number of molecular units of the structure is shown in an inconsistent way. Some structures contain full biological unit coordinates data, whereas some others have information about only one DNA strand. Although PDB now provides a biological unit database for its entries, maintaining a local mirror of the same is not well supported. Owing to this difficulty the second option of the webserver i.e. calculation of conformational parameters from a PDB code should be used with care. If there is a discrepancy between biological unit and general coordinate files of PDB, users are encouraged to download a biological unit coordinate file from PDB and directly submit it to the server.
NMR structure models
PDB has structures determined by using NMR and the entry for these structures usually consists of several models. In our webserver, we use only the first model of the PDB file for calculating conformational properties. All subsequent models are ignored.
Non-standard nucleic acid bases
Force field used in this webserver was derived from a non-redundant dataset of protein–DNA complexes in PDB (9). Only the four standard bases, namely, A, C, G and T were used to generate this force field. Thus, the server does not calculate the conformational energy or Z-scores for DNA whose sequence contains an identification code other than these four standard bases. A warning message will be displayed in such cases.
Sequence size limit
Calculation of Z-scores using the above method requires calculation of base–amino acid contacts for the target structure and also energy for a large set of random sequences. The number of random DNA sequences needed for generating a converging solution of the Z-scores increases rapidly with increasing the sequence length considered. We have therefore limited the DNA sequence length to 50 nucleic acid bases in the current version of the server.
Acknowledgments
This work is supported in part by Grants-in-Aid for Scientific Research 16014219, 17017030, 17054031 (A.S.) and 16014226 (H.K.) from Ministry of Education, Culture, Sports, Science and Technology in Japan. One of the authors (M.J.A.-B.) would like to acknowledge the Japanese Society for Promotion of Science (JSPS) for supporting him for this research. Funding to pay the Open Access publication charges for this article was provided by the above grants to A.S.
Conflict of interest statement. None declared.
REFERENCES
- 1.Sarai A., Kono H. Protein–DNA recognition patterns and predictions. Ann. Rev. Biophys. Biomol. Struct. 2005;34:379–398. doi: 10.1146/annurev.biophys.34.040204.144537. [DOI] [PubMed] [Google Scholar]
- 2.Olson W.K., Gorin A.A., Lu X.J., Hock L.M., Zhurkin V.B. DNA sequence-dependent deformability deduced from protein–DNA crystal complexes. Proc. Natl Acad. Sci. USA. 1998;95:11163–11168. doi: 10.1073/pnas.95.19.11163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Otwinowski Z., Schevitz R.W., Zhang R.G., Lawson C.L., Joachimiak A., Marmorstein R.Q., Luisi B.F., Sigler P.B. Crystal structure of the trp repressor/operator complex at atomic resolution. Nature. 1988;33:321–329. doi: 10.1038/335321a0. [DOI] [PubMed] [Google Scholar]
- 4.Drew H.R., Travers A.A. Structural junctions in DNA: the influence of flanking sequence on nuclease digestion specificities. Nucleic Acids Res. 1985;13:4445–4467. doi: 10.1093/nar/13.12.4445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gromiha M., Siebers J.G., Selvaraj S., Kono H., Sarai A. Intermolecular and intramolecular readout mechanisms in protein–DNA recognition. J. Mol. Biol. 2004;337:285–294. doi: 10.1016/j.jmb.2004.01.033. [DOI] [PubMed] [Google Scholar]
- 6.Sarai A., Mazur J., Nussinov R., Jernighan R.L. Sequence dependence of DNA conformational flexibility. Biochemistry. 1989;28:7842–7849. doi: 10.1021/bi00445a046. [DOI] [PubMed] [Google Scholar]
- 7.Hogan M.E., Austin R.H. Importance of DNA stiffness in protein–DNA binding specificity. Nature. 1987;329:263–266. doi: 10.1038/329263a0. [DOI] [PubMed] [Google Scholar]
- 8.Kono H., Sarai A. Structured-based prediction of DNA target sites by regulatory proteins. Proteins. 1999;35:114–131. [PubMed] [Google Scholar]
- 9.Selvaraj S., Kono H., Sarai A. Specificity of protein–DNA recognition revealed by structure-based potentials: symmetric/asymmetric and cognate/non-cognate binding. J. Mol. Biol. 2002;322:907–915. doi: 10.1016/s0022-2836(02)00846-x. [DOI] [PubMed] [Google Scholar]
- 10.Pichierri F., Aida M., Gromiha M.M., Sarai A. Free energy maps of base–amino acid interaction for protein–DNA recognition. J. Am. Chem. Soc. 1999;121:6152–6157. [Google Scholar]
- 11.Yoshida T., Nishimura T., Aida M., Pichierri F., Gromiha M.M., Sarai A. Evaluation of free energy landscape for base-amino acid interactions using ab initio force field and extensive sampling. Biopolymers. 2002;61:84–95. doi: 10.1002/1097-0282(2001)61:1<84::AID-BIP10045>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- 12.Packer M.J., Dauncey M.P., Hunter C.A. Sequence-dependent DNA structure: tetranucleotide conformational maps. J Mol. Biol. 2000;295:85–103. doi: 10.1006/jmbi.1999.3237. [DOI] [PubMed] [Google Scholar]
- 13.Sponer J., Leszczynski J., Hobza P. Hydrogen bonding and stacking of DNA bases: a review of quantum-chemical ab initio studies. J Biomol. Struct. Dyn. 1996;14:117–135. doi: 10.1080/07391102.1996.10508935. [DOI] [PubMed] [Google Scholar]
- 14.Piacenza M., Grimme S. Systematic quantum chemical study of DNA-base tautomers. J. Comput. Chem. 2004;25:83–99. doi: 10.1002/jcc.10365. [DOI] [PubMed] [Google Scholar]
- 15.Paillard G., Lavery R. Analyzing protein–DNA recognition mechanisms. Structure. 2004;12:113–122. doi: 10.1016/j.str.2003.11.022. [DOI] [PubMed] [Google Scholar]
- 16.Beveridge D.L., Barreiro G., Byun K.S., Case D.A., Cheatham T.E., III, Dixit S.B., Giudice E., Lankas F., Lavery R., Maddocks J.H., et al. Molecular dynamics simulations of the 136 unique tetranucleotide sequences of DNA oligonucleotides. I. Research design and results on d(CpG) steps. Biophys. J. 2004;87:3799–3813. doi: 10.1529/biophysj.104.045252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Araúzo-Bravo M.J., Fujii S., Kono H., Ahmad S., Sarai A. Sequence-dependent conformational energy of DNA derived from molecular dynamics simulations: toward understanding the indirect readout mechanism in protein–DNA recognition. J. Am. Chem. Soc. 2005;127:16074–16089. doi: 10.1021/ja053241l. [DOI] [PubMed] [Google Scholar]
- 18.Sarai A., Siebers J., Selvaraj S., Gromiha M.M., Kono H. Integration of bioinformatics and computational biology to understand protein–DNA recognition mechanism. J. Bioinform. Comput. Biol. 2005;3:169–183. doi: 10.1142/s0219720005000965. [DOI] [PubMed] [Google Scholar]
- 19.Lu X.J., Olson W.K. 3DNA: a software package for the analysis,rebuilding and visualization of three-dimensional nucleic acid structures. Nucleic Acids Res. 2003;31:5108–5121. doi: 10.1093/nar/gkg680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Olson W.K., Bansal M., Burley S.K., Dickerson R.E., Gerstein M., Harvey E.C., Heinemann U., Lu X.J., Neidle S., Shakked Z., et al. A standard reference frame for the description of nucleic acid base-pair geometry. J. Mol. Biol. 2001;313:229–237. doi: 10.1006/jmbi.2001.4987. [DOI] [PubMed] [Google Scholar]
- 21.Dickerson R.E., Bansal M., Calladine C.R., Diekmann S., Hunter W.N., Kennard O., Kitzing E., Lavery R., Nelson H.C.M., Olson W.K., et al. Definitions and nomenclature of nucleic acid structure parameters. Nucleic Acids Res. 1989;17:1797–1803. doi: 10.1093/nar/17.5.1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Berman H.M., Westbrook J., Feng Z., Gilliland G., Bhat T.N., Weissig H., Shindyalov I.N., Bourne P.E. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]