Abstract
Variable order Markov models and variable order Bayesian trees have been proposed for the recognition of transcription factor binding sites, and it could be demonstrated that they outperform traditional models, such as position weight matrices, Markov models and Bayesian trees. We develop a web server for the recognition of DNA binding sites based on variable order Markov models and variable order Bayesian trees offering the following functionality: (i) given datasets with annotated binding sites and genomic background sequences, variable order Markov models and variable order Bayesian trees can be trained; (ii) given a set of trained models, putative DNA binding sites can be predicted in a given set of genomic sequences and (iii) given a dataset with annotated binding sites and a dataset with genomic background sequences, cross-validation experiments for different model combinations with different parameter settings can be performed. Several of the offered services are computationally demanding, such as genome-wide predictions of DNA binding sites in mammalian genomes or sets of 104-fold cross-validation experiments for different model combinations based on problem-specific data sets. In order to execute these jobs, and in order to serve multiple users at the same time, the web server is attached to a Linux cluster with 150 processors. VOMBAT is available at http://pdw-24.ipk-gatersleben.de:8080/VOMBAT/.
INTRODUCTION
One important and interesting problem in genome research is the prediction of transcription factor binding sites (TFBSs). Binding of transcription factors to their DNA binding sites in the regulatory region of a gene is a prerequisite for its activation or repression. The combinatorial presence of TFBSs controls gene regulation at least partially, and the prediction of TFBSs is of importance for unraveling the underlying molecular mechanisms.
Wet-lab experiments allow the identification of TFBSs, but these experiments are expensive and time consuming. Computational methods, which are often less accurate but easy to conduct with available resources, are a welcome complementation to wet-lab experiments. Many TFBS prediction algorithms employ statistical models for scoring a given sequence as TFBS or non-TFBS. For these algorithms, the chosen family of statistical models is of importance for the performance of prediction.
The family of Markov models (1–3) is chosen in many TFBS prediction algorithms as well as for a variety of other classification problems. Well-known examples are the position weight matrix (PWM) model (4), which is an inhomogeneous Markov model of order 0, and the weight array matrix (WAM) model (5), which is an inhomogeneous Markov model of order 1. From a statistical point of view, Markov models employ assumptions about the statistical independence of nucleotides at different positions of the binding site.
The strongest independence assumption is realized by the PWM model, where each position is assumed to be statistically independent of all other positions. As indicated e.g. in (6), it is an open question whether this strong independence assumption is reasonable in view of recent results indicating the presence of statistical dependences between positions (7,8). Markov models of higher order take into account statistical dependences from previous positions, called the context, where the length of the context is equal to the order of the Markov model.
Despite the often unjustified independence assumption, PWM models are often found to outperform Markov models of higher order in the prediction of TFBSs. This is probably caused by the rather limited amount of training data available from experimentally verified TFBSs. As the number of model parameters grows exponentially with the order of the Markov model, Markov models of higher order tend to be over-fitted, resulting in poor performance.
One possibility to circumvent this problem is provided by variable order Markov models, which do not require the contexts to be of a fixed length, but allow contexts with variable lengths (9–12). The power of variable order Markov models stems from the freedom to include only those contexts into the model for which there are strong statistical dependences.
Another possibility to capture statistical dependences between non-adjacent positions without the burden of increasing the number of model parameters exponentially are provided by Bayesian trees (BTs) (13–16). In BTs the contexts are not restricted to the previous positions within the binding site, and it could be demonstrated that BTs outperform PWM models and WAM models in the prediction of splice sites (17,18) and TFBSs (6,19).
As an extension of variable order Markov models and BTs, we introduce variable order BTs (19). The VOMBAT web server is designed to train variable order Markov models and variable order BTs on user-supplied data and to apply the resulting models to the prediction of TFBSs. In addition, the VOMBAT web server provides a cross-validation platform, allowing advanced users to compare different model combinations with different parameter settings on problem-specific datasets. The VOMBAT web server is primarily designed for the prediction of TFBSs, but applicable to other types of fixed-length motifs as well.
ALGORITHM
The prediction of TFBSs is based on one statistical model for TFBSs and one for non-TFBS sequences constituting the background. Each statistical model assigns a probability P(x1⋯xL) to a given DNA sequence x1⋯xL of L nucleotides. For any value of M, 1 ≤ M ≤ L, P(x1⋯xL) can be decomposed according to
Markov models of order M assume that the conditional probabilities Pl(xl∣x1⋯xl−1) do not depend on all previous nucleotides x1⋯xl−1, but only on the M previous nucleotides xl−M⋯xl−1, which are called the context of position l. Hence, a Markov model of order M is defined by
If the conditional probabilities Pl(xl∣xl−M⋯xl−1) are identical at all positions, the Markov model is called homogeneous, otherwise it is called inhomogeneous.
In contrast to a Markov model of order 1, where the conditional probabilities at position l depend only on the previous nucleotide xl−1, a BT allows that the conditional probabilities at position l may depend on the nucleotide xPa(l) at a possibly non-adjacent position Pa(l), which is called the parent of position l. In a BT, arbitrarily remote positions may be chosen as parents as long as no cycles between positions are induced. This implies that there is one position, called the root position, which must not depend on any other position, and that the statistical dependences of a BT may be graphically represented by a rooted tree, where the positions are represented by nodes and the statistical dependences are represented by edges (13–16). The probability distribution defined by a BT with root position r decomposes as
In the VOMBAT web server, the so-called motif model Pmotif (used for modeling TFBSs) and the so-called background model Pbg (used for modeling background DNA sequences) can be chosen by the user. An inhomogeneous Markov model or a BT should be chosen if statistical dependences are assumed to vary from position to position within the sequence. This is typically the case for TFBSs or other motifs. A homogeneous Markov model should be chosen if statistical dependences are assumed to be the same at all positions within the sequence. This is a reasonable assumption for background sequences. The appropriate order of a Markov model depends on the expected range of statistical dependences and on the amount of available training data.
For Markov models of fixed order, the number of model parameters grows exponentially with the order, resulting in a sharp transition from under-fitted to over-fitted models. Variable order Markov models were developed to circumvent this sharp transition (9–12). The idea of variable order Markov models is to shorten the context in those cases where extending the context does not yield ‘enough’ statistical dependences. Mathematically this is formulated by measuring to which degree the conditional probabilities change when extending or reducing the context.
The Kullback–Leibler divergence (20) is used as a measure of change of the conditional probability distributions. It measures the degree of dissimilarity of the conditional probability distribution given the extended context and the conditional probability distribution given the reduced context. The Kullback–Leibler divergence is always non-negative, but small positive values of the Kullback–Leibler divergence do not indicate significant statistical dependences. The threshold above which the Kullback–Leibler divergences are considered important can be set by an external parameter, c, called pruning constant.
For c = 0, any extension of the context is considered important, and the resulting variable order Markov model becomes a traditional Markov model of order M. For c → ∞, any extensions of the context is considered unimportant, and the resulting variable order Markov model becomes a traditional Markov model of order 0, i.e. a PWM model. As c grows from 0 to ∞, more and more contexts are considered unimportant, and the resulting variable order Markov models become smaller and smaller, interpolating between a Markov model of order M and a PWM model. More detail and other aspects can be found in (19).
The parameters of the conditional probabilities are estimated from user-supplied training data. VOMBAT uses a maximum likelihood estimator with optional pseudo counts, which can be specified by the user, for smoothing the conditional probability distributions and to compensate for zero occurrences of nucleotides and contexts. The motif model Pmotif is learned from a user-supplied training set of TFBSs, and the background model Pbg is learned from a user-supplied training set of background sequences.
After training Pmotif and Pbg, VOMBAT can be used to compute genome-wide predictions of the learned motif in a set of user-supplied sequences. For each position of a sliding window of length L, the log-likelihood ratio of the oligonucleotide y1⋯yL occurring at that position is computed, and y1⋯yL is predicted as TFBS if the log-likelihood ratio is greater than a given threshold T, i.e. if
Specific knowledge, such as the number of TFBSs expected in the genome, can be taken into account to adjust the threshold T to achieve TFBS predictions with the desired balance of sensitivity and specificity.
For advanced users, VOMBAT allows a cross-validation of the prediction accuracy of a user-defined combination of Pmotif and Pbg based on a user-supplied set of TFBSs and a user-supplied set of background sequences. From each of the datasets, 10% of the sequences are randomly excluded, and the models Pmotif and Pbg are trained on the remaining 90%. Subsequently, both models are used for the prediction of the withheld data, and the sensitivity given a user-defined value of the specificity is computed. This procedure is repeated k times, where k can be specified by the user, and the mean sensitivity and its standard error are reported.
VOMBAT is based on a variant of the three-tier architecture, where the presentation layer, the management layer and the execution framework are logically separated and physically located on different servers. The web front-end of VOMBAT is based on standard technologies like JavaServer Faces for the web forms and Servlets for displaying images and providing downloads of results. The user-supplied parameters and input files are stored in a mySQL-database. This database is queried by the execution framework. If the execution framework detects a job to be executed, it requests the necessary parameters and input files and submits the corresponding job to the scheduler of the attached Linux cluster. After the job is finished, the results are written to the database and can be displayed by the web front-end, which loads numerical results, model descriptions and images from the result-tables of the mySQL-database.
INPUT AND OUTPUT
Imagine a user interested in a genome-wide prediction of binding sites of the transcription factor SP1 in GC-poor upstream regions of human RefSeq (21) genes. If the user were interested in a prediction based on pre-trained PWM models stored in the Transfac database (22), he could use the TFBS prediction program MATCH™ (3). If the user were interested in a prediction based on variable order Markov models or variable order BTs, he could use the VOMBAT web server.
The two main functions of the VOMBAT web server are training variable order Markov models or variable order BTs based on user-supplied sequences and predicting putative TFBSs based on user-supplied variable order Markov models or variable order BTs. The training function allows the user to choose an optimal combination of a motif model and a background model and train these models on problem-specific datasets. For example, the user might choose a variable order BT as motif model and a variable order Markov model of order 5 as background model, he might choose that subset of SP1 binding sites available from Transfac that are located in GC-poor promoters as motif dataset, and he might choose a representative set of GC-poor promoter regions as background dataset.
Training a model
For training a model from user-supplied input data, the user first selects ‘Train a model from data’ from the selection of tasks. The subsequently displayed form requests the necessary parameters. The user can enter a comment on the model to be trained in order to make it easier to identify the corresponding results in a list of all results. The rest of the parameters defines the type of model to be trained as well as the pruning constant and the pseudo count. The default value of the pseudo count is 1, and we recommend the user to always specify a pseudo count greater than 0 to compensate for zero occurrences.
The subsequent three parameters define the type of model to be trained. The first parameter is the initial order of the model. The second parameter defines if the model to be trained is homogeneous or inhomogeneous. If the model is defined to be inhomogeneous, the user can select between a Markov model and a BT as a third parameter. Based on (6,19) and based on systematic cross-validation analyses of different model combinations applied to different sets of TFBSs and background sequences, we generally recommend to use a BT as motif model and a homogeneous Markov model with an initial order of at least 3 as background model.
The following two parameters are data specific. The user-supplied input file to train an inhomogeneous model must consist of aligned sequences of identical length, separated by line breaks. For homogeneous models the length of the sequences may differ. The VOMBAT web server is designed for, but not restricted to, the prediction of motifs in DNA sequences. Hence, it is possible to specify the alphabet of the sequences. In case of DNA sequences the alphabet is ‘ACGT.’ As a last option the user may select a checkbox that determines if a graphical representation of the trained model shall be displayed as Supplementary Data.
If the job of training a model could be successfully submitted to the scheduler of the attached Linux cluster, the subsequently displayed page reports ‘Jobs success,’ and VOMBAT gives the user the possibility to go either to the list of tasks for starting another job or to the job overview.
The job overview displays a table of all jobs with the following columns: the time at which the job was started, the comment that was entered by the user, the type of the job (‘Train’, ‘Classify’, ‘CrossVal’), the current state of the job, a button for the already available results and a button to stop a running job. The possible states of a job are ‘START’ for jobs still waiting for execution, ‘RUNNING’ for jobs currently running on the Linux cluster, ‘STOP’ for jobs marked to be stopped, ‘STOPPED’ for stopped jobs and ‘FINISHED’ for successfully executed jobs. If the state of a job is ‘FINISHED,’ the user can click on the button ‘View results…’ in the same row to inspect the results of this job.
The results of training a model are a link to an XML representation of the trained model, which can be saved and used as input for one of the other tasks presented in the following. If the user selected to obtain a graphical representation of the trained model, this representation is also displayed as a series of images, which may be saved using the corresponding browser command.
Predicting putative transcription factor binding sites
After a motif model and a background model have been trained and saved, they can be combined to predict putative TFBSs in a user-supplied set of input sequences. For this purpose, the user selects ‘Classify data’ from the list of tasks. The first input field of the subsequently displayed form, which is shown in Figure 1, allows to enter a user-defined comment on the job. Next, the user uploads two files containing the XML representations of the desired motif and background models, which have been trained using the ‘Train a model from data’ task described above. The classification threshold must be specified, and the file containing the set of sequences in which the motif is to be predicted must be uploaded.
Figure 1.
The form requesting the parameters for a TFBS prediction.
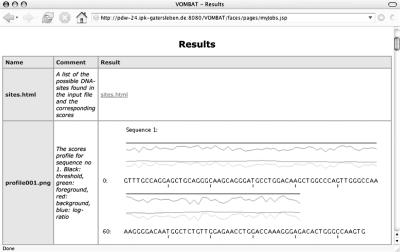
After a classification job has been started, the user can monitor the current state of the job in the job overview of VOMBAT. If the job is finished, the predictions can be inspected by clicking on the corresponding ‘View results…’button. The subsequently displayed result page is presented in Figure 2. The HTML file contains a textual description of the putative motifs, which contains for each predicted motif its position, its sequence, its probabilities returned by the two models, and the corresponding log-likelihood ratio. Additionally, a profile of the log-likelihoods and the log-likelihood ratio is plotted for each sequence of the input file. The chosen classification threshold is also plotted as a reference for the log-likelihood ratio, allowing the user to judge the influence of the classification threshold on the prediction results at a glance.
Figure 2.
The results of a TFBS prediction. The list of putative TFBSs are displayed as a link to ‘sites.html,’ and the profiles for the sequences are plotted.
Cross-validation
As an auxiliary function for advanced users, VOMBAT provides a platform for cross-validation analyses of different combinations of user-defined models based on problem-specific data. Selecting ‘Cross validation’ from the list of tasks brings up the cross-validation input form. The first input field allows to enter a user-defined comment on the cross-validation job. Next, the number of iterations of the cross-validation job can be entered. A higher number of iterations produces more reliable results. On the other hand, the cross-validation job becomes more time-consuming.
Two files containing XML representations of the motif model and the background model must be uploaded by the user. These files can be obtained by a standard training on the datasets to be used for cross-validation.
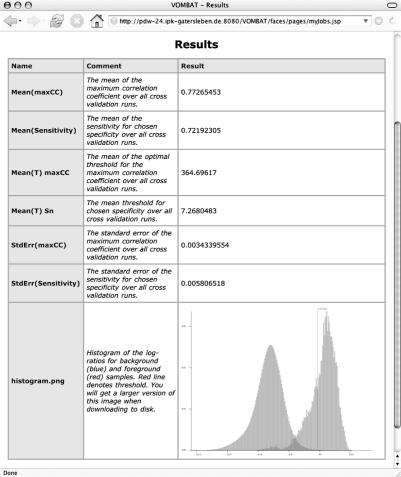
Next, the user must specify the desired specificity for which the mean sensitivity is to be computed. The last option in the form allows to plot a histogram of the log-likelihood ratios for the motif sequences and background sequences, which allows further analyses of the cross-validation results, such as judging the effect of a shifted threshold or inspecting the separation of the samples of both classes.
The cross-validation results are also listed in the job overview. The presented results are the mean (and standard error) of the sensitivity for the fixed specificity entered in the cross-validation form, the mean (and standard error) of the maximum correlation coefficient, and the corresponding thresholds and optionally the histogram of the log-likelihood ratios as shown in Figure 3.
Figure 3.
The results of a cross-validation.
The following prototypical example illustrates how the three functions of VOMBAT may be utilized for the prediction of putative TFBSs. Recall the user interested in a genome-wide prediction of SP1 binding sites. Typically, the user has no a priori knowledge of which model combination and which model parameters could provide an accurate prediction of these binding sites in GC-poor upstream regions. Hence, he would start a series of different cross-validation experiments for different model combinations and different parameter settings using the cross-validation function of VOMBAT.
After having established an optimal model combination (including optimal parameters) for his specificity requirements and based on his problem-specific datasets, e.g. based on already annotated SP1 binding sites from Transfac as motif dataset and representative GC-poor upstream regions as background dataset, he would then use these models to run the training and prediction functions of VOMBAT as outlined above.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR online.
Acknowledgments
We thank André Gohr for implementing many of the algorithms, Martin Oertel for valuable discussions, and the German Ministry of Education and Research (BMBF Grant No. 0312706A/D) for financial support. Funding to pay the Open Access publication charges for this article was provided by IPK Gatersleben.
Conflict of interest statement. None declared.
REFERENCES
- 1.Fickett J., Hatzigeorgiou A. Eukaryotic promoter recognition. Genome Res. 1997;7:861–878. doi: 10.1101/gr.7.9.861. [DOI] [PubMed] [Google Scholar]
- 2.Salzberg S. A method for identifying splice sites, translational start sites in eukaryotic mRNA. CABIOS. 1997;13:365–376. doi: 10.1093/bioinformatics/13.4.365. [DOI] [PubMed] [Google Scholar]
- 3.Kel A.E., Gossling E., Reuter I., Cheremushkin E., Kel-Margoulis O.V., Wingender E. MATCH™: a tool for searching transcription factor binding sites in DNA sequences. Nucleic Acids Res. 2003;31:3576–3579. doi: 10.1093/nar/gkg585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Staden R. Computer methods to locate signals in nucleic acid sequences. Nucleic Acids Res. 1984;12:505–519. doi: 10.1093/nar/12.1part2.505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang M.Q., Marr T.G. A weight array method for splicing signal analysis. Comput. Appl. Biosci. 1993;9:499–509. doi: 10.1093/bioinformatics/9.5.499. [DOI] [PubMed] [Google Scholar]
- 6.Barash Y., Elidan G., Friedman N., Kaplan T. Modeling dependencies in protein-dna binding sites. Proceedings of the Annual International Conference on Research in Computational Molecular Biology (RECOMB); 2003. pp. 28–37. [Google Scholar]
- 7.Benos P.V., Lapedes A.S., Fields D.S., Stormo G.D. Samie: statistical algorithm for modeling interaction energies. Pacific Symposium on Biocomputing; 2001. pp. 115–126. ACM, NY. [DOI] [PubMed] [Google Scholar]
- 8.Bulyk M.L., Johnson P.L.F., Church G.M. Nucleotides of transcription factor binding sites exert interdependent effects on the binding affinities of transcription factors. Nucleic Acids Res. 2002;30:1255–1261. doi: 10.1093/nar/30.5.1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rissanen J. A universal data compression system. IEEE Transactions on Information Theory. 1983;29:656–664. [Google Scholar]
- 10.Ron D., Singer Y., Tishby N. The power of amnesia: learning probabilistic automata with variable memory length. Mach. Learn. 1996;25:117–149. [Google Scholar]
- 11.Buhlmann P., Wyner A.J. Variable length markov chains. Ann. Statist. 1999;27:480–513. [Google Scholar]
- 12.Bejerano G., Yona G. Variations on probabilistic suffix trees: statistical modeling and prediction of protein families. Bioinformatics. 2001;17:23–43. doi: 10.1093/bioinformatics/17.1.23. [DOI] [PubMed] [Google Scholar]
- 13.Chow C.K., Liu C.N. Approximating discrete probability distributions with dependence trees. IEEE Transactions on Information Theory. 1968;14:462–467. [Google Scholar]
- 14.Pearl J. Probabilistic Reasoning in Intelligent Systems: Networks of plausible inference. San Mateo, CA: Morgan Kaufmann; 1988. [Google Scholar]
- 15.Buntine W. Theory refinement on bayesian networks. Proceedings of Seventh Conference on Uncertainty in Artificial Intelligence; Los Angeles, CA. Morgan Kaufmann; 1991. pp. 52–60. [Google Scholar]
- 16.Heckerman D., Geiger D., Chickering D.M. Learning bayesian networks. the combination of knowledge and statistical data. Machine Learn. 1995;20:197–243. [Google Scholar]
- 17.Cai D., Delcher A., Kao B., Kasif S. Modeling splice sites with bayes networks. Bioinformatics. 2000;16:152–158. doi: 10.1093/bioinformatics/16.2.152. [DOI] [PubMed] [Google Scholar]
- 18.Castelo R., Guigo R. Splice site identification by idlbns. Bioinformatics. 2004;20:i69–i76. doi: 10.1093/bioinformatics/bth932. [DOI] [PubMed] [Google Scholar]
- 19.Ben-Gal I., Shani A., Gohr A., Grau J., Arviv S., Shmilovici A., Posch S., Grosse I. Identification of transcription factor binding sites with variable-order bayesian networks. Bioinformatics. 2005;21:2657–2666. doi: 10.1093/bioinformatics/bti410. [DOI] [PubMed] [Google Scholar]
- 20.Kullback S., Leibler R.A. On information and sufficiency. Ann. Math. Statist. 1951;22:79–86. [Google Scholar]
- 21.Pruitt K.D., Tatusova T., Maglott D.R. Ncbi reference sequence (refseq): a curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res. 2005;33:D501–D504. doi: 10.1093/nar/gki025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wingender E., Chen X., Fricke E., Geffers R., Hehl R., Liebich I., Krull M., Matys V., Michael H., Ohnhäuser R., et al. The transfac system on gene expression regulation. Nucleic Acids Res. 2001;29:281–283. doi: 10.1093/nar/29.1.281. [DOI] [PMC free article] [PubMed] [Google Scholar]