Abstract
Seventy-eight human and environmental strains of Salmonella enterica subsp. enterica serovar Typhimurium, as well as 18 isolates of other Salmonella serovars and 6 isolates of Escherichia coli, were subjected to a novel variable number of tandem repeats (VNTR)-based fingerprinting method that showed high discrimination and reproducibility for typing serovar Typhimurium isolates. The method is based on capillary separation of PCR products from fluorescence-labeled VNTR in the serovar Typhimurium genome. The serovar Typhimurium isolates displayed 54 VNTR patterns, and the VNTR assay correctly identified strains from a well-characterized outbreak. Among 37 serovar Typhimurium phage type DT104 isolates, 28 distinct VNTR patterns were found. This VNTR-based method is fast and suitable for complete automation. Our VNTR-based method was capable of high discrimination within the homogeneous serovar Typhimurium DT104 phage type and can be used to trace outbreaks and to monitor DT104 as well as other phage types. The VNTR assay was compared to XbaI pulsed-field gel electrophoresis, amplified fragment length polymorphism analysis, integron-cassette profiles and gene PCR of intI1, qacEΔ1, sulI1, and floR. The VNTR assay showed greatly improved resolution compared to all other tested methods in this study.
The class of repetitive DNA named variable number of tandem repeats (VNTR) is, in general, a source of genetic polymorphism in humans. VNTR have been extensively studied in humans and can consist of several hundreds to several thousands of base pairs of DNA in head-to-tail repetition of short sequence motifs of about 10 to 100 bp (20). The spontaneous mutation rate to new alleles is sufficiently high to be measured in human pedigrees, and VNTR can possess several alleles (3, 19) and be implicated in human disease (28). VNTR have gained interests in prokaryotes when complete bacterial genomes were sequenced. Several bacterial strains have multiple VNTR in their genome, and in some instances, they have been adopted for typing purposes. A VNTR-based assay has recently been developed for typing the pathogen Bacillus anthracis using eight VNTR loci. Characterization of 426 B. anthracis isolates with this method gave 89 distinct genotypes (22). Polymorphic VNTR regions which can be used for typing purposes have been identified and tested in Yersinia pestis (1, 23), Francisella tularensis (21), Mycobacterium tuberculosis (10-12, 37, 40, 41), Xylella fastidiosa (6), Haemophilus influenzae (43, 44, 49), and Bacillus anthracis (2, 8, 18, 22, 38). The VNTR-based typing approach is promising in all of these strains. A database of tandem repeats in several completely sequenced genomes is also available (26). VNTR appear to contain a high level of polymorphism, which gives a high discriminatory capacity.
Salmonella enterica serovar Typhimurium is the only salmonella serovar that causes recurrent domestic infections in Norway. The method of choice for typing serovar Typhimurium as a means of source identification, outbreak investigation, and phylogenic studies is, and has been, macrorestriction with pulsed-field gel electrophoresis (PFGE) separation of the fragments. PFGE has good discriminatory power and has proven highly useful and reliable in outbreak situations (1). The PFGE method is, however, quite labor-intensive, and the resulting electrophoretic patterns can be difficult to compare between different runs. The discriminatory power of the PFGE method has, however, been challenged with the introduction and spread of serovar Typhimurium definite-type 104 (DT104). This phage type displays a high degree of homogeneity, and epidemiologically related, as well as unrelated, strains display identical PFGE profiles. Several reports show the difficulty of typing DT104 isolates with PFGE, and no other method has surfaced as a preferred typing alternative. In a recent report, serovar Typhimurium DT104 isolates from 1985, 1990, and 1995 were shown to have indistinguishable PFGE and integron profiles, which supports the idea of a clonal relationship between recent and historical isolates (34). In an Irish study 122 human and animal serovar Typhimurium DT104 isolates as well as 6 DT104b isolates from human and animal products were typed by PFGE using the three enzymes SpeI, SfiI, and XbaI (31). This study showed that 81.2% of the isolates still displayed identical genetic fingerprints (31). This homogeneity challenges the usefulness of PFGE in outbreak investigations caused by serovar Typhimurium DT104.
Amplified-fragment length polymorphism (AFLP) analysis is a DNA fingerprinting method based on restriction cutting of DNA and stringent PCR amplification of the resulting fragments. This method is faster than PFGE but also seems to give a low discrimination on DT104 isolates (25). To try to address this typing problem, the sequenced serovar Typhimurium LT2 genome (29) was analyzed to identify and pick regions containing repeated DNA.
We present here a VNTR-based typing assay for the important food pathogen serovar Typhimurium with special emphasis on DT104. This method shows a much higher discrimination than PFGE and is easier to use as well as being considerably faster. The assay was robust and gave identical results on repeated tries. In addition, the resulting banding patterns were easy to interpret and treat digitally in the BioNumerics software package. This method is suitable for full automation since its main steps involve setting up and pooling PCRs with subsequent capillary electrophoresis, where the first step easily can be performed by a robotic system.
MATERIALS AND METHODS
Bacterial strains.
In all 78 isolates of serovar Typhimurium, 6 isolates of serovar Enteritidis, 3 isolates of serovar Albany, 2 isolates each of serovar Panama and serovar Paratyphi B variant Java, as well as 1 isolate each of Salmonella serovars Shwarzengrund, a [4,5,12:i:-] monophasic variant, Anatum, Blockley, Saintpaul, and Typhi together with 6 Escherichia coli strains were obtained from the strain collection at the National Reference Laboratory for Enteropathogenic Bacteria at the Norwegian Institute of Public Health, Oslo. Thirty-seven of the isolates were serovar Typhimurium DT104 (Table 1) and the remaining were isolates containing phage types U302, DT204, DT120, and DT104A as well as isolates not reacting to the phages or not phage typed.
TABLE 1.
Characteristics of DT104 isolates used in this study
| Isolate | Yr | Origin | Integron(s)a (bp) | Resistanceb | Amplification productc
|
|||
|---|---|---|---|---|---|---|---|---|
| int1 | qacEΔ1 | sul1 | floR | |||||
| 3919/00 | 2000 | Norway | 1,000, 1,200 | ACSSuT | + | + | + | + |
| 4536/00 | 2000 | Not known | 1,000, 1,200 | ACSSuT | + | + | + | + |
| 2755/97 | 1997 | Norway | 1,000, 1,200 | ACSuT | + | + | + | + |
| 3104/97 | 1997 | Philippines | 1,000, 1,200 | ACSSuT | + | + | + | + |
| 1252/99 | 1999 | Norway | 1,000, 1,200 | ACSSuT | + | + | + | + |
| 164/99 | 1999 | Denmark | 1,000, 1,200 | ACSSuT | + | + | + | + |
| 1804/98 | 1998 | England | 1,000, 1,200 | ACSSuT | + | + | + | + |
| 1805/96 | 1996 | Spain | 1,000, 1,200 | ACST | + | + | − | + |
| 1227/98 | 1998 | Norway | 1,000, 1,200 | ACST | + | + | − | + |
| 700/98 | 1998 | Norway | 1,000, 1,200 | ACST | − | + | − | + |
| 2663/96 | 1996 | Cyprus | 1,000, 1,200 | ACSSuT | + | + | + | + |
| 6347/00 | 2000 | Spain | 1,000, 1,200 | ACSSuT | − | + | + | + |
| 1961/96 | 1996 | Norway | 1,000, 1,200 | ACSSuT | + | + | + | + |
| 11325 | 2001 | Sweden | 1,000, 1,200 | ACSSuT | + | + | + | + |
| 10399 | 2001 | Australia | 1,000, 1,200 | ACSSuT | + | + | + | + |
| 10111242 | 2001 | Norway | 1,000, 1,200 | ACSSuT | + | + | + | + |
| 10381 | 2001 | Australia | 1,000, 1,200 | ACSSuT | + | + | + | + |
| 3990/00 | 2000 | Mediterranean region | 1,000, 1,200 | ACSSuT | + | + | + | + |
| 10382 | 2001 | Australia | 1,000, 1,200 | ACSSuT | + | + | + | + |
| 10385 | 2001 | Australia | 1,000, 1,200 | ACSSuT | + | + | + | + |
| 10400 | 2001 | Australia | 1,000, 1,200 | ACSSuT | + | + | + | + |
| 1011095 | 2001 | Norway | 1,000, 1,200 | ACSSuT | + | + | + | + |
| 3544/00 | 2000 | Spain | 1,000, 1,200 | ACSSuT | + | + | + | + |
| 3592/00 | 2000 | Spain | 1,000, 1,200 | ACSSuT | + | + | + | + |
| 11327 | 2001 | Sweden | 1,000, 1,200 | ACSSuT | + | + | + | + |
| 8709/00 | 2000 | Norway | 1,000, 1,200 | ACSSuT | + | + | + | + |
| 10402 | 1999 | Australia | 1,000 | SSu | + | + | + | + |
| 10110953 | 2001 | Norway | 1,000, 1,200 | ACSSuT | + | + | + | + |
| 10110636 | 2001 | Germany | 1,000, 1,200 | ACSSuT | + | + | + | + |
| 10110637 | 2001 | Germany | 1,000, 1,200 | ACSSuT | + | + | + | + |
| 10111243 | 2001 | Norway | 1,000, 1,200 | ACSSuT | + | + | + | + |
| 2755/97 | 1997 | Norway | 1,000, 1,200 | ACSSuT | + | + | + | + |
| 6723/00 | 2000 | Cyprus | 1,000, 1,200 | ACSSuT | + | + | + | + |
| 6861/00 | 2000 | Cyprus | 1,000, 1,200 | ACSSuT | + | + | + | + |
| 2658/99 | 1999 | Cyprus | 1,000, 1,200 | ACSSuT | + | + | + | + |
| 3087/99 | 1999 | Sweden | 1,000, 1,200 | ACSSuT | + | + | + | + |
Sizes of products amplified with primers directed at the integron variable region (gene cassettes) (27). The 1,000-bp product corresponds with band 1 in Fig. 2, and the 1,200-bp product corresponds with band 2 in Fig. 2.
Abbreviations for resistance patterns: A, ampicillin; C, chloramphenicol; S, streptomycin; Su, sulfadiazine; T, tetracycline.
+, presence of an amplification product from the primers directed at the indicated gene.
Testing for resistance to antibiotics.
Antimicrobial susceptibility was tested by a tablet diffusion method according to the manufacturer's guidelines (Rosco Diagnostics, Taastrup, Denmark). Neo-Sensitabs are antibiotic containing tablets 9 mm in diameter that are standardized according to susceptibility testing standardization groups in several countries, including Norway, Holland, Sweden, Denmark, United Kingdom, Germany, and France. In short, an inoculum of dense but not completely confluent growing bacteria is dropped on an agar plate (PDM-II susceptibility test media; Biodisk AB, Solna, Sweden) and distributed evenly across the surface. The plate is then allowed to dry before the tablets are placed onto the agar surface and incubated for 24 h before inhibition zones are recorded. The isolates were screened for resistance to ampicillin, ciprofloxacin, tetracycline, chloramphenicol, nalidixic acid, trimethoprim-sulfamethoxazole, sulfadiazine, streptomycin, and gentamicin.
VNTR typing.
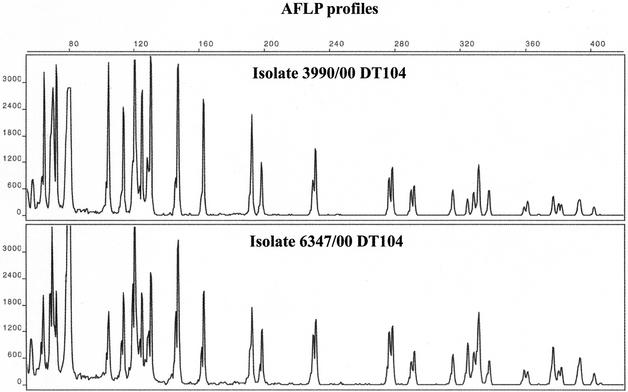
The genomic sequence of serovar Typhimurium LT2 (29) was analyzed using GeneQuest (DNASTAR) and Repeat Finder (4) software. Regions containing direct repeats in six coding regions and two intergenic regions with repeat motifs from 6 to 189 bp in length were further studied, and a subset was tested for their usefulness in a set of well-characterized serovar Typhimurium isolates, both DT104 and non-DT104 strains. Several strains from a known serovar Typhimurium outbreak in Norway in 1999 were also included. The repeat areas were located, and both software programs reported multiple repeated regions within the serovar Typhimurium LT2 genome. An initial selection of eight direct tandem repeats were chosen after comparing the type of repeats, homogeneity, and copy number. The repeats can be shown in Table 2. Six of the repeats were located inside open reading frames reported by the GeneQuest program (Table 2). The actual repeats with leader and tailing sequence of 150 to 200 bp were used to design specific PCR primers (Table 3). Care was taken to match both annealing temperature and sizes of the produced PCR amplicons to make a convenient set for PCR and capillary electrophoresis separation. The PrimerSelect (DNASTAR) software automatically calculated the primer annealing temperatures and analyzed how the different primers could interact in multiplex PCR. The upper primers for all the eight VNTR regions were labeled with 5′-carboxyfluorescein (FAM). The different primer sets were tested against each other in several combinations. It was clear that multiplexing of most of the primer sets was possible except STTR6 and STTR8, which had lower annealing temperatures. However, when the different multiplexing mixes were performed and tested by both agarose gel electrophoresis and capillary electrophoresis, it was evident that the best result was obtained by combining only two primers at a time and pooling the PCR products before electrophoresis. The primer combinations from loci STTR1 and STTR2; STTR3 and STTR4; and STTR5 and STTR7 were annealed at 58°C, while STTR6 and STTR8 were annealed at 55°C. The PCRs were carried out in 20-μl reaction mixtures on a Perkin-Elmer GeneAmp 9700 PCR system (ABI Biosystems) with AmpliTaq polymerase (ABI Biosystems) and 1× buffer (containing 1.5 mM MgCl2) supplied with enzyme. The temperature profile was as follows: 94°C denaturation for 5 min; 30 cycles of 94°C for 30 s, annealing temperature (given above) for 30 s, and 72°C for 50 s; and finally a 7-min extension step at 72°C. Ten microliters of each of the PCR products, resulting from the different multiplexed primer sets amplified from the same strain, was pooled together into a single tube and dried in a vacuum concentrator (Speedvac; Thermo Savant, Holbrook, N.Y.) before resuspension in 20 μl of water. Thus, each pooled tube now contained a mixture of all the amplified VNTR for one isolate. One microliter of the resuspended solution from each sample was then used for capillary electrophoresis on an ABI-310 Genetic Analyzer (ABI Biosystems, Foster City, Calif.) with POP4-polymer and Genescan TAMRA-500 or TAMRA-2500 as internal standard in each sample (ABI Biosystems). The resulting electropherograms of the pooled amplicons showed clear and easily interpretable banding patterns (Fig. 1). The electropherograms were imported into the BioNumerics software package, and a phylogenetic tree was constructed using Dice coefficients and cluster analysis with the unweighted pair group method with arithmetic averages from the ABI trace files.
TABLE 2.
Selected VNTR loci in the serovar Typhimurium LT2 genome (GenBank accession no. AE006468)
| Locus | Repeat length (bp) | No. of repeats (bp) | Gene | % GC content | Location in LT2 (nt) | Functiona |
|---|---|---|---|---|---|---|
| STTR1 | 21 | 15 | tolA | 66.8 | 815055-815395 | Putative membrane-spanning protein |
| STTR2 | 60 | 10 | sspH2 | 58.9 | 2341937-2342552 | Signaling protein (29) |
| STTR3 | 33 | 10 | bigA | 65.9 | 3629542-3629900 | Putative surface-exposed virulence protein |
| STTR4 | 189 | 5 | shdA | 61.2 | 2630632-2631588 | Putative Peyer's patch colonization and shredding factor |
| STTR5 | 6 | 13 | yohM | 66.2 | 3184543-3184622 | Putative inner membrane protein |
| STTR6 | 6 | 13 | 65.9 | 2730867-2730948 | Prophage related (9, 29, 39) | |
| STTR7 | 39 | 8 | ftsK | 69.8 | 1039570-1039883 | Putative cell division protein |
| STTR8 | 116 | 4 | 49.1 | 3414113-3414676 | No information |
TABLE 3.
Primers for PCR amplification of selected VNTR loci in serovar Typhimurium LT2 (GenBank accession no. AE006468)
| Locus | Primer | Primer sequence | Product size (bp) | Optimal annealing temp (°C) |
|---|---|---|---|---|
| STTR1 | STTR1-U | 5′-FAM-CAGCAGTACAACCGTCAGCAGGAT | 770 | 62.3 |
| STTR1-L | 5′-GCCCCACCGTTAGCGCCCGATGTA | |||
| STTR2 | STTR2-U | 5′-FAM-GGGTTCCCTTCCAGATTTACG | 711 | 58.9 |
| STTR2-L | 5′-TTTACCCGCGCATATTACCACACT | |||
| STTR3 | STTR3-U | 5′-FAM-CCCCCTAAGCCCGATAATGG | 490 | 61.8 |
| STTR3-L | 5′-TGACGCCGTTGCTGAAGGTAATAA | |||
| STTR4 | STTR4-U | 5′-FAM-GGCGCCGCTATGGGTGGTGA | 1,138 | 60.7 |
| STTR4-L | 5′-CAAAAGCGGTAGCGATGAGC | |||
| STTR5 | STTR5-U | 5′-FAM-ATGGCGAGGCGAGCAGCAGT | 259 | 58.9 |
| STTR5-L | 5′-GGTCAGGCCGAATAGCAGGAT | |||
| STTR6 | STTR6-U | 5′-FAM-TCGGGCATGCGTTGAAA | 342 | 54.3 |
| STTR6-L | 5′-CTGGTGGGGAGAATGACTGG | |||
| STTR7 | STTR7-U | 5′-FAM-CGCGCAGCCGTTCTCACT | 594 | 60.9 |
| STTR7-L | 5′-TGTTCCAGCGCAAAGGTATCTA | |||
| STTR8 | STTR8-U | 5′-FAM-TTATGTCACCACGGCTGTCAAT | 925 | 57.2 |
| STTR8-L | 5′-AAGGCCAAATAGGGGTTCATAAGG |
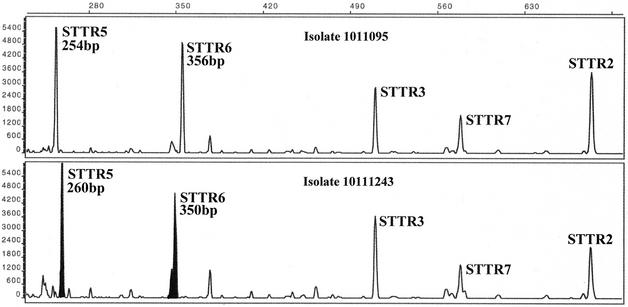
FIG. 1.
Electropherogram from pooled capillary electrophoresis run of FAM-labeled products from the STTR5, STTR6, STTR3, STTR7, and STTR2 VNTR loci of the 1011095 DT104 isolate (upper panel) and the 10111243 DT104 isolate (lower panel). Vertical scale is relative fluorescence. Horizontal scale is size in base pairs. The STTR5 VNTR is 1 repeat unit longer (+6 bp) in 10111243 than in 1011095, and the STTR6 VNTR is 1 repeat unit shorter (−6 bp) in 10111243 than in 1011095.
PFGE.
A standard XbaI macrorestriction fragment was subjected to PFGE as previously detailed (16). The DNA fragments were separated in 1% SeaKem GTG agarose, (FMC, Rockland, Maine) with 0.25× modified Tris-borate-EDTA buffer for 22 h at 350 V and 12°C with pulse times from 5 to 40 s using a Beckman (Fullerton, Calif.) Gene Line II.
Integron PCR.
For the DT104 isolates PCRs with primers for amplification of integrons (5′-GGCATCCAAGCAGCAAG and 5′-AAGCAGACTTGACCTGA) (27) were performed, as well as PCRs with primers for the integron integrase gene intI1 (5′-GCCTTGCTGTTCTTCTACGG and 5′-GATGCCTGCTTGTTCTACGG) (27), the sulfonamide resistance gene sulI (5′-CTTCGATGAGAGCCGGCGGC and 5′-GCAAGGCGGAAACCCGCGCC) (35), the florfenicol resistance gene floR (5′-ACCCGCCCTCTGGATCAAGTCAAG and 5′-CAAATCACGGGCCACGCTGTATC), and the disinfectant resistance gene qacEΔ1 (5′-ATCGCAATAGTTGGCGAAGT and 5′-CAAGCTTTTGCCCATGAAGC) (35). The temperature profile was 94°C denaturation for 5 min; 25 cycles of 94°C for 30 s, 55°C for 30 s and 72°C for 30 s; and then a 7-min extension step at 72°C on a Perkin-Elmer GeneAmp PCR system 9700 (ABI Biosystems) for the intI1, qacEΔ1, and integron cassette primers. The sulI and floR primers had the same PCR profile but with annealing at 60°C. The PCR products were separated on a 1% GTG agarose gel and visualized using a GelDoc-2000 (Bio-Rad, Hercules, Calif.) after ethidium bromide staining. An example of the results from the primers directed at the integron variable region can be seen in Fig. 2.
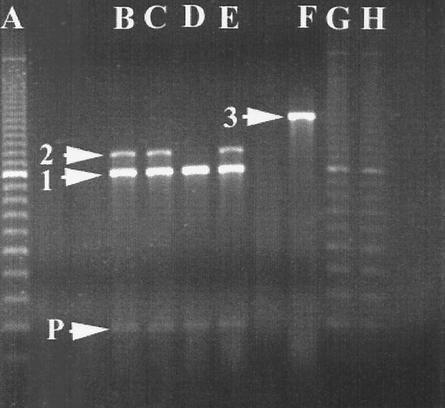
FIG. 2.
Example of PCR amplification of integrons using primers directed to the integron variable region (gene cassette region). Lane A, 100-bp ladder; lane B, the integron profile of serovar Typhimurium DT104 isolate 10111243; lane C, the integron profile of serovar Typhimurium DT104 isolate 1805/96; lane D, the integron profile of serovar Typhimurium DT104 isolate 10402; lane E, the integron profile of serovar Typhimurium DT104 isolate 11327; lane F, a control PCR with a strain known to harbor a 1,600-bp integron; lanes G and H, 100-bp ladder. Bands: 1, 1,000 bp; 2, 1,200 bp; 3, 1,600 bp; P, 200-bp PCR artifact caused by primer mispairing.
AFLP analysis.
Genomic DNA was extracted using a commercial kit (Easy-DNA; Invitrogen BV, Leek, The Netherlands). We used a modification of the AFLP protocol first described by Vos et al. (46). The combination which gave the fingerprint patterns reported here was the EcoRI(0) + BamHI(0) primer combination. This resulted in fingerprints with bands up to about 500 bp. The EcoRI adapters were as follows: 5′-AAT TGG TAC GCA GTC TAC GAG-3′ and 5′-CTC GTA GAC TGC GTA −3′. The BamHI adapters were as follows: 5′-GAC GAT GAG TCC TGA-3′ and 5′-GAT CCT CAG GAC TCA TCG TC-3′. The BamHI PCR primer was 5′ labeled with the dye FAM and had the following sequence: 5′-FAM-GAC GAT GAG TCC TGA GGA TC-3′. The EcoRI primer had the sequence 5′-TCG TAG ACT GCG TAC CAA TT-3′. For the restriction cutting and ligation, 500 ng of genomic DNA was incubated at 37°C for 3 h in a 20-μl solution containing 1× NEB buffer for EcoRI (New England Biolabs, Beverly, Mass.) with 10 U each of EcoRI and BamHI (New England Biolabs). After 3 h the digested fragments were cleaned with the MinElute Reaction Cleanup kit (QIAGEN, Hilden, Germany) and resuspended in 10 μl with NEB buffer. To the resuspended cleaned fragments, 1× T4 DNA ligase buffer, 25 pmol of annealed BamHI adapters, 25 pmol of annealed EcoRI adapters, and 5 U of T4 DNA ligase (New England Biolabs) were added. Water was then added to make a 20-μl final ligation mixture. This ligation mixture was incubated for 3 h at 16°C. Four microliters of the ligation mixture was used in a 20-μl PCR mix containing 10 pmol of primers for EcoRI and BamHI, a 2 mM concentration of each deoxynucleoside triphosphate, and 0.4 U of Taq polymerase (Sigma, St. Louis, Mo.) in 1× Taq buffer supplied with enzyme. The PCR was carried out on a Perkin-Elmer GeneAmp PCR system 9700 (ABI Biosystems). The temperature profile was as follows: 95°C denaturation for 5 min; 10 cycles of 94°C for 30 s, 60°C for 30 s, and 72°C for 45 s; then 30 cycles of 94°C for 30 s, 56°C for 30 s, 72°C for 1 min; and finally a 5-min extension step at 72°C. Two microliters of the PCR products was taken out for capillary electrophoresis on an ABI-310 Genetic Analyzer (ABI Biosystems) with POP4-polymer and Genescan TAMRA-500 as internal standard in each sample (ABI Biosystems).
RESULTS
A total of 102 isolates, including 37 isolates of DT104, were examined using eight VNTR loci. The resulting electropherograms from the VNTR-typing assay were clear (Fig. 1) and easily imported into BioNumerics for analysis. It was evident after running the electrophoresis from 50 to 120 min, and after looking at the degree of discrimination provided, that the level of strain discrimination presented here could be achieved by a 50-min electrophoresis time and inclusion of five of the eight examined VNTR. The results presented in this report are based on the STTR5, STTR6, STTR3, STTR7, and the STTR2 VNTR loci (Fig. 1). The VNTR loci displayed a wide range of polymorphisms in the serovar Typhimurium strains, with the 6-bp repeats as the most polymorphous. The STTR5 locus in the yohM gene displayed 11 different alleles among the serovar Typhimurium isolates. The STTR6 locus had eight alleles. The STTR3 locus in the bigA gene had five different alleles. For the STTR7 locus only two alleles could be seen, with one allele represented only once across all serovar Typhimurium isolates. No size variation in the STTR2 locus located in the sspH2 gene could be seen. By not being present in all isolates, the STTR7 and STTR2 loci still gave useful typing information. The VNTR typing assay showed a high degree of discrimination of serovar Typhimurium strains.
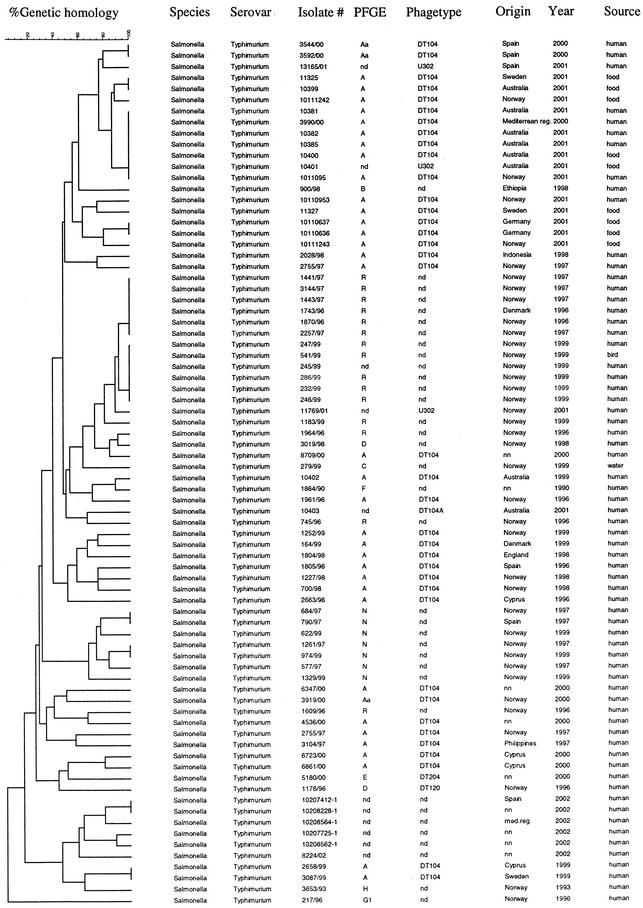
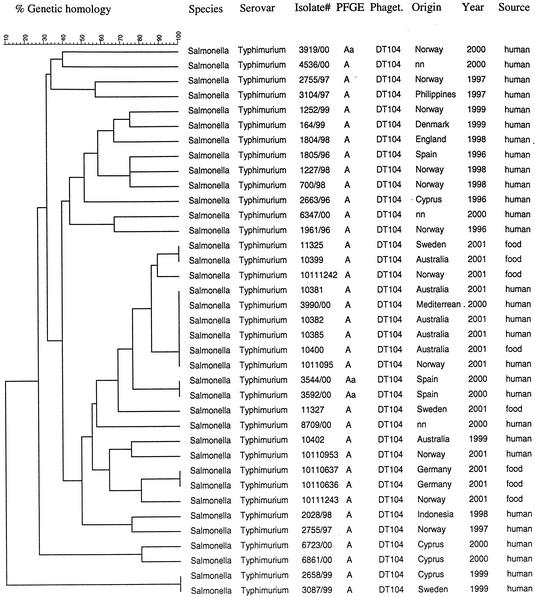
The VNTR assay grouped the isolates 247/99, 541/99, 286/99, 245/99, 232/99, and 246/99 together with identical fingerprints (Fig. 3). These isolates are from a well-defined serovar Typhimurium outbreak in western Norway in 1999. The isolate 541/99 was the source strain originating from a seagull, and 247/99, 286/99, 232/99, 245/99, and 246/99 are isolates from individual patients. The cluster with isolates 1441/97, 3144/97 1443/97, 1743/96 1870/96, and 2257/97 is made up of strains from the same geographic region as the 1999 outbreak in western Norway. They all share a similar PFGE pattern (R) with the outbreak strains and would not be separated from the outbreak strains by the use of PFGE alone. The VNTR assay showed that these isolates are indeed clonal but distinct from the isolates from the outbreak. The human and food strains 11325, 10399, 10381, 10385, 10382, 1011095, 10400, 10111242, 10110637, 10110636, 10111243, 11327, 10110953, and 10403 were isolated in several countries suspected of being part of a worldwide outbreak currently under investigation. XbaI PFGE of these isolates gave an identical genetic fingerprint (A) for all strains (10401 and 10403 were not typed by PFGE). The VNTR assay, however, showed that the DT104 isolates 10381, 10385, 10382, 1011095, 10400; the U302 isolate 10401; and the 3990/00 strain displayed identical VNTR profiles (Fig. 3). The 3990/00 strain was not isolated as a suspected outbreak strain, but the VNTR fingerprinting showed that this isolate was identical to six other isolates collected as outbreak strains. The other clusters of the suspected outbreak strains consisted of 10110637 with 10110636 and 11325 with 10399. The remaining strains in this suspected outbreak all gave different VNTR profiles. The identical DT104 isolates 3544/00 and 3592/00 were both isolates from patients infected on the Spanish island of Tenerife, who became sick only 2 days apart, suggesting a common source. The isolates 10207412-1 and 102082288-1 had identical VNTR profiles, but due to incomplete data on isolate 102082288-1 this relationship was only confirmed by the VNTR assay. All the remaining isolates had distinct VNTR profiles, giving a total of 54 VNTR profiles (Fig. 3). When looking only at the DT104 isolates (Fig. 4), the VNTR assay distinguished 28 different profiles, while the XbaI PFGE could only distinguish one main profile (A) and a closely related profile (Aa) showing a one-band difference (Fig. 5).
FIG. 3.
Dendrogram of all the VNTR-typed serovar Typhimurium strains. Abbreviations: nd, phage typing or PFGE not done; nn, origin of strain not certain.
FIG. 4.
Dendrogram of the VNTR-typed serovar Typhimurium DT104 strains only. nn, origin of strain not certain.
FIG. 5.
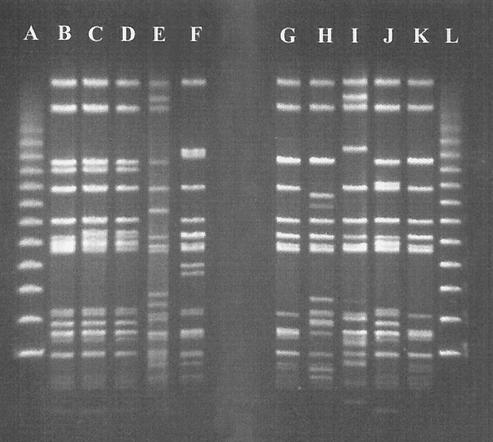
XbaI PFGE gel with the following profiles: lane A, λ size marker; lane B, A profile; lane C, Aa profile from 3544/00; lane D, Aa profile from 3919/00; lane E, B profile; lane F, F profile; lane G, D profile from 3019/98; lane H, E profile; lane I, C profile; lane J, H profile; lane K, D profile from 1176/96; lane L, λ size marker.
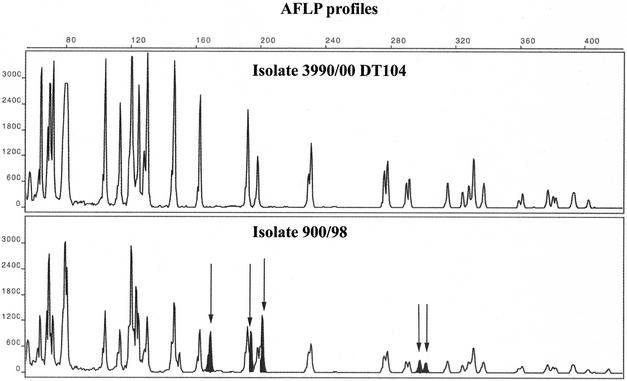
The BamHI+EcoRI AFLP assay did not discriminate between any of the 37 serovar Typhimurium DT104 isolates and showed the same pattern for all DT104 strains (Fig. 6); however, it managed to distinguish the DT104 AFLP pattern from some of the non-DT104 serovar Typhimurium isolates (Fig. 7).
FIG. 6.
AFLP electropherograms of two serovar Typhimurium DT104 isolates. The two strains have identical AFLP patterns, and this pattern is shared by all the DT104 isolates in this study.
FIG. 7.
AFLP electropherograms of two serovar Typhimurium isolates, one DT104 (upper panel) and one not DT104 but multiresistant to antibiotics (lower panel). Some variations between the two patterns can be seen. The variation is marked with filled peaks and arrows in the lower panel.
Our VNTR assay did not, however, group the DT104 isolates as a distinct cluster, but spread the DT104 patterns out among the other serovar Typhimurium isolates. It also gave identical fingerprints between a U302 strain (10401) and a cluster of DT104 isolates (Fig. 3).
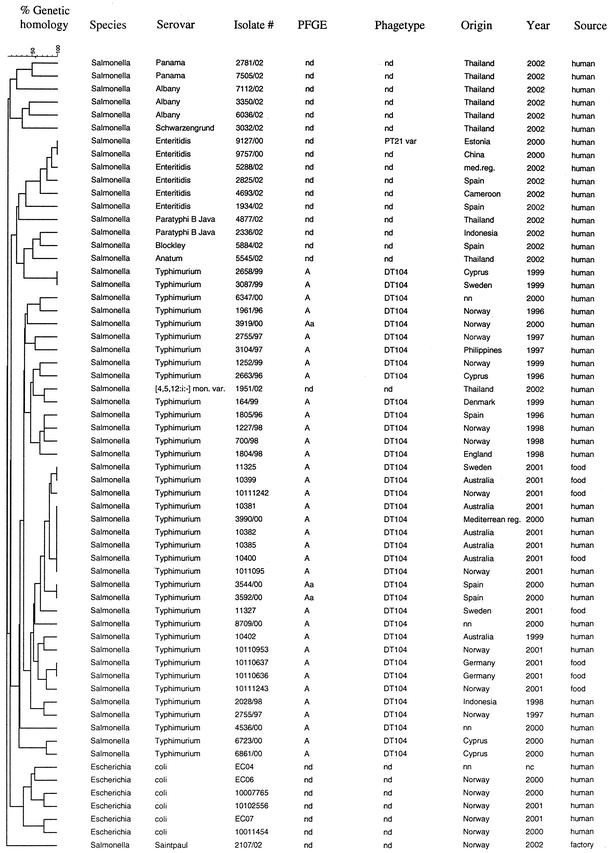
Since the genes containing the VNTR repeats were orthologs of genes in E. coli and also believed to be present in other serovars of Salmonella, we additionally performed the VNTR assay on a selection of Salmonella serovars different from Typhimurium as well as on six E. coli isolates. In Fig. 8, the serovar Typhimurium DT104 isolates are shown together with the E. coli and the non-Typhimurium strains. This was done to determine whether other Salmonella serovars or E. coli could display identical profiles to any of the serovar Typhimurium isolates. The result showed that the E. coli and the non-Typhimurium serovars had distinct VNTR patterns from the serovar Typhimurium strains (Fig. 8), although two of the DT104 isolates (2658/99 and 3087/99) clustered with the non-Typhimurium strains.
FIG. 8.
Comparison of the serovar Typhimurium strains with isolates of other Salmonella serovars and isolates of E. coli. Abbreviations: nd, phagetyping not done; nn, origin of strain not certain; nc, year of isolation not certain.
DISCUSSION
One of the most common strains of the genus Salmonella currently isolated from humans is the multidrug-resistant serovar Typhimurium DT104 (14, 30, 33, 42). Serovar Typhimurium DT104 is an important zoonotic pathogen affecting primarily cattle; however, species such as cats, horses, pigs, and sheep can also be affected (45, 48; E. J. Threlfall, F. J. Angulo, and P. G. Wall, Letter, Vet. Rec. 142:255, 1998; P. G. Wall, E. J. Threllfall, L. R. Ward, and B. Rowe, Letter, Lancet 348:471, 1996). The hospitalization and mortality rates are higher for DT104 than other Salmonella serotypes (47), and excess mortality associated with drug-resistant serovar Typhimurium strains was noted in a recent study (17). DT104 has also been shown to give a more severe illness in calves than what is usual for Salmonella (5). This makes the ability to characterize and subtype serovar Typhimurium DT104 isolates important for tracing infections and investigating outbreaks. The genetic homogeneity of DT104 is, unfortunately, such that subtyping by PFGE displays low resolution. The resolution can be improved by using several different restriction endonucleases, but it still appears low and the time used is also increased. We decided to see if a new typing scheme, based on VNTR, would increase the discriminatory power for genetic typing of serovar Typhimurium isolates with emphasis on DT104. The nature of tandem repeats make them intrinsically prone to a higher degree of genetic variability, and in a genetically homogenous pathogen like DT104, it is possible that only such repeated regions have had the time to display the polymorphism needed for useful typing purposes. A number of studies have shown that tandem repeats are likely to be an important source of informative markers for the identification of pathogenic bacteria, even when these pathogens have recently emerged and are highly homogeneous. VNTR typing assays have been used successfully with bioterrorism-related bacteria and also in strains where typing by PFGE has been of limited value (15). Tandem repeats may play a role in adapting pathogens to their hosts (7, 13) and are additionally means for rapid phase variations in bacteria (32).
The main finding of this study was the high discrimination of the serovar Typhimurium DT104 bacterial isolates. All the DT104 strains used in this study were typed by PFGE displaying identical patterns (A), except for the isolates 3919/00, 3544/00, and 3592/00, which were internally identical and displayed a small one-band difference (Aa pattern) from our main DT104 XbaI PFGE profile (Fig. 5). The DT104 isolates were also checked for the occurrence of integrons (Table 1). All the DT104 isolates were additionally typed by AFLP using the EcoRI and the BamHI restriction endonucleases. The AFLP results confirmed the genetic homogeneity displayed by PFGE but also grouped the PFGE A-pattern isolates with the Aa-pattern isolates giving all DT104 identical fingerprints. This is in accordance with a previous study showing that AFLP with the EcoRI and MseI restriction endonucleases cannot discriminate within DT104 isolates (25). Thus, for the majority of our isolates we had identical patterns by PFGE and AFLP as well as identically sized integrons.
The results from our VNTR assay showed that it correctly identified and grouped together strains from a well-characterized serovar Typhimurium outbreak in western Norway in 1999 and that it could discriminate between isolates of the DT104 phage type that were identical by PFGE, AFLP, and integron pattern analyses (Fig. 3 and 4).
Thirteen of the non-DT104 serovar Typhimurium isolates displayed the PFGE R profile, where five of these (247/99, 541/99, 286/99, 232/99, and 246/99) were outbreak strains and eight were not related to that outbreak. Isolate 245/99 was not PFGE typed but was isolated as part of the 1999 outbreak. The VNTR assay, unlike PFGE, managed to discriminate the outbreak strains from the unrelated PFGE R profiles (Fig. 3). The PFGE R strains grouped into two main clusters with 100% identity between the strains internally in each cluster. One cluster was the 1999 outbreak and the other consisted of isolates from the same geographic region as the outbreak, with the exception of strain 1743/96, which was isolated from a patient after traveling to Denmark. This isolate was, however, submitted from a hospital in the same geographic region as the other isolates in this cluster, indicating that this patient might also have been infected in the same region.
The VNTR assay grouped the serovar Typhimurium U302 isolate 10401 together with a cluster of DT104 strains containing isolates from patients and suspected food. The 10401 U302 strain was isolated from the same location and the same food source as the 10400 DT104 isolate in the same cluster, both suspected as outbreak sources. This makes the location of the U302 strain in this cluster likely from an epidemiological point of view (Fig. 3). This might indicate that the phage typing scheme used for discrimination between Salmonella isolates can in some instances lead to separation of strains that are otherwise genetically identical. It has been argued that some DT104 strains have developed an altered susceptibility to some of the typing phages and thus display different phage types (24). The typing phages have, additionally, been shown to undergo recombination when they are passed through a host containing a related prophage, creating recombinants which may have different plating properties (36).
We observed that the VNTR-based typing method did not separate the multiresistant strains into clusters separate from the nonresistant strains. Thus, clustering by this VNTR assay is independent of whether or not the isolates are resistant to antibiotics. None of the multiresistant DT104 isolates were identical to any of the tested susceptible isolates; however, the clustering of the U302 isolates showed that this method placed them both among multiresistant and susceptible isolates. The VNTR loci used in this study are believed to be in genes coding for membrane-associated proteins and virulence proteins which have no known association with functions related to antibiotic resistance. This may explain the lack of coclustering of antibiotic-resistant isolates.
In conclusion we propose this VNTR-based typing assay for rapid typing and high-level discrimination of serovar Typhimurium isolates, with a special emphasis on its usefulness for fingerprinting DT104 isolates. This method can be adapted to automation and will give relatively rapid answers in an outbreak situation involving the genetically homogenous and multiresistant DT104 phage type. At the time of writing this appears to be the only DNA-based typing method with high enough discrimination to fingerprint serovar Typhimurium DT104 at the resolution needed for outbreak investigations.
Acknowledgments
The staff of the National Salmonella Reference Laboratory is gratefully acknowledged for technical assistance, and librarian Winifred J. Larsen is acknowledged for assistance with English grammar and orthography.
REFERENCES
- 1.Adair, D. M., P. L. Worsham, K. K. Hill, A. M. Klevytska, P. J. Jackson, A. M. Friedlander, and P. Keim. 2000. Diversity in a variable-number tandem repeat from Yersinia pestis. J. Clin. Microbiol. 38:1516-1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Andersen, G. L., J. M. Simchock, and K. H. Wilson. 1996. Identification of a region of genetic variability among Bacillus anthracis strains and related species. J. Bacteriol. 178:377-384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Armour, J. A., and A. J. Jeffreys. 1992. Biology and applications of human minisatellite loci. Curr. Opin. Genet. Dev. 2:850-856. [DOI] [PubMed] [Google Scholar]
- 4.Benson, G. 1999. Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Res. 27:573-580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carlson, S. A., W. C. Stoffregen, and S. R. Bolin. 2002. Abomasitis associated with multiple antibiotic resistant Salmonella enterica serotype Typhimurium phagetype DT104. Vet. Microbiol. 85:233-240. [DOI] [PubMed] [Google Scholar]
- 6.Coletta-Filho, H. D., M. A. Takita, A. A. de Souza, C. I. Aguilar-Vildoso, and M. A. Machado. 2001. Differentiation of strains of Xylella fastidiosa by a variable number of tandem repeat analysis. Appl. Environ. Microbiol. 67:4091-4095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.de La Fuente, J., J. C. Garcia-Garcia, E. F. Blouin, S. D. Rodriguez, M. A. Garcia, and K. M. Kocan. 2001. Evolution and function of tandem repeats in the major surface protein 1a of the ehrlichial pathogen Anaplasma marginale. Anim. Health Res. Rev. 2:163-173. [PubMed] [Google Scholar]
- 8.Enserink, M. 2001. Taking anthrax's genetic fingerprints. Science 294:1810-1812. [DOI] [PubMed] [Google Scholar]
- 9.Figueroa-Bossi, N., S. Uzzau, D. Maloriol, and L. Bossi. 2001. Variable assortment of prophages provides a transferable repertoire of pathogenic determinants in Salmonella. Mol. Microbiol. 39:260-271. [DOI] [PubMed] [Google Scholar]
- 10.Filliol, I., S. Ferdinand, L. Negroni, C. Sola, and N. Rastogi. 2000. Molecular typing of Mycobacterium tuberculosis based on variable number of tandem DNA repeats used alone and in association with spoligotyping. J. Clin. Microbiol. 38:2520-2524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Frothingham, R. 1995. Differentiation of strains in Mycobacterium tuberculosis complex by DNA sequence polymorphisms, including rapid identification of M. bovis BCG. J. Clin. Microbiol. 33:840-844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Frothingham, R., and W. A. Meeker-O'Connell. 1998. Genetic diversity in the Mycobacterium tuberculosis complex based on variable numbers of tandem DNA repeats. Microbiology 144:1189-1196. [DOI] [PubMed]
- 13.Ge, Z., and D. E. Taylor. 1999. Contributions of genome sequencing to understanding the biology of Helicobacter pylori. Annu. Rev. Microbiol. 53:353-387. [DOI] [PubMed] [Google Scholar]
- 14.Glynn, M. K., C. Bopp, W. Dewitt, P. Dabney, M. Mokhtar, and F. J. Angulo. 1998. Emergence of multidrug-resistant Salmonella enterica serotype typhimurium DT104 infections in the United States. N. Engl. J. Med. 338:1333-1338. [DOI] [PubMed] [Google Scholar]
- 15.Harrell, L. J., G. L. Andersen, and K. H. Wilson. 1995. Genetic variability of Bacillus anthracis and related species. J. Clin. Microbiol. 33:1847-1850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Heir, E., B. A. Lindstedt, T. Vardund, Y. Wasteson, and G. Kapperud. 2000. Genomic fingerprinting of shigatoxin-producing Escherichia coli (STEC) strains: comparison of pulsed-field gel electrophoresis (PFGE) and fluorescent amplified-fragment-length polymorphism (FAFLP). Epidemiol. Infect. 125:537-548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Helms, M., P. Vastrup, P. Gerner-Smidt, and M. lbak. 2002. Excess mortality associated with antimicrobial drug-resistant salmonella typhimurium. Emerg. Infect. Dis. 8:490-495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jackson, P. J., E. A. Walthers, A. S. Kalif, K. L. Richmond, D. M. Adair, K. K. Hill, C. R. Kuske, G. L. Andersen, K. H. Wilson, M. Hugh-Jones, and P. Keim. 1997. Characterization of the variable-number tandem repeats in vrrA from different Bacillus anthracis isolates. Appl. Environ. Microbiol. 63:1400-1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jeffreys, A. J., N. J. Royle, V. Wilson, and Z. Wong. 1988. Spontaneous mutation rates to new length alleles at tandem-repetitive hypervariable loci in human DNA. Nature 332:278-281. [DOI] [PubMed] [Google Scholar]
- 20.Jeffreys, A. J., V. Wilson, and S. L. Thein. 1985. Hypervariable ′minisatellite' regions in human DNA. Nature 314:67-73. [DOI] [PubMed] [Google Scholar]
- 21.Johansson, A., I. Goransson, P. Larsson, and A. Sjostedt. 2001. Extensive allelic variation among Francisella tularensis strains in a short-sequence tandem repeat region. J. Clin. Microbiol. 39:3140-3146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Keim, P., L. B. Price, A. M. Klevytska, K. L. Smith, J. M. Schupp, R. Okinaka, P. J. Jackson, and M. E. Hugh-Jones. 2000. Multiple-locus variable-number tandem repeat analysis reveals genetic relationships within Bacillus anthracis. J. Bacteriol. 182:2928-2936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Klevytska, A. M., L. B. Price, J. M. Schupp, P. L. Worsham, J. Wong, and P. Keim. 2001. Identification and characterization of variable-number tandem repeats in the Yersinia pestis genome. J. Clin. Microbiol. 39:3179-3185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lawson, A. J., M. U. Dassama, L. R. Ward, and E. J. Threlfall. 2002. Multiply resistant (MR) Salmonella enterica serotype Typhimurium DT 12 and DT 120: a case of MR DT 104 in disguise? Emerg. Infect. Dis. 8:434-436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Leegaard, T. M., D. A. Caugant, L. O. Frøholm, E. A. Høiby, and J. Lassen. 2000. Emerging antibiotic resistance in Salmonella typhimurium in Norway. Epidemiol. Infect. 125:473-480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Le Fleche, P., Y. Hauck, L. Onteniente, A. Prieur, F. Denoeud, V. Ramisse, P. Sylvestre, G. Benson, F. Ramisse, and G. Vergnaud. 2001. A tandem repeats database for bacterial genomes: application to the genotyping of Yersinia pestis and Bacillus anthracis. BMC Microbiol. 1:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Levesque, C., L. Piche, C. Larose, and P. H. Roy. 1995. PCR mapping of integrons reveals several novel combinations of resistance genes. Antimicrob. Agents Chemother. 39:185-191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lindstedt, B. A., D. Ryberg, and A. Haugen. 1997. Rare alleles at different VNTR loci among lung-cancer patients with microsatellite instability in tumours. Int. J. Cancer 70:412-415. [DOI] [PubMed] [Google Scholar]
- 29.McClelland, M., K. E. Sanderson, J. Spieth, S. W. Clifton, P. Latreille, L. Courtney, S. Porwollik, J. Ali, M. Dante, F. Du, S. Hou, D. Layman, S. Leonard, C. Nguyen, K. Scott, A. Holmes, N. Grewal, E. Mulvaney, E. Ryan, H. Sun, L. Florea, W. Miller, T. Stoneking, M. Nhan, R. Waterston, and R. K. Wilson. 2001. Complete genome sequence of Salmonella enterica serovar Typhimurium LT2. Nature 413:852-856. [DOI] [PubMed] [Google Scholar]
- 30.Mead, P. S., L. Slutsker, V. Dietz, L. F. McCaig, J. S. Bresee, C. Shapiro, P. M. Griffin, and R. V. Tauxe. 1999. Food-related illness and death in the United States. Emerg. Infect. Dis. 5:607-625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Murphy, T. M., E. McNamara, M. Hill, N. Rooney, J. Barry, J. Egan, A. O'Connell, J. O'Loughlin, and S. McFaddyen. 2001. Epidemiological studies of human and animal Salmonella typhimurium DT104 and DT104b isolates in Ireland. Epidemiol. Infect. 126:3-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Peak, I. R., M. P. Jennings, D. W. Hood, M. Bisercic, and E. R. Moxon. 1996. Tetrameric repeat units associated with virulence factor phase variation in Haemophilus also occur in Neisseria spp. and Moraxella catarrhalis. FEMS Microbiol. Lett. 137:109-114. [DOI] [PubMed] [Google Scholar]
- 33.Poppe, C., N. Smart, R. Khakhria, W. Johnson, J. Spika, and J. Prescott. 1998. Salmonella typhimurium DT104: a virulent and drug-resistant pathogen. Can. Vet. J. 39:559-565. [PMC free article] [PubMed] [Google Scholar]
- 34.Ribot, E. M., R. K. Wierzba, F. J. Angulo, and T. J. Barrett. 2002. Salmonella enterica serotype Typhimurium DT104 Isolated from Humans, United States, 1985, 1990, and 1995. Emerg. Infect. Dis. 8:387-391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sandvang, D., F. M. Aarestrup, and L. B. Jensen. 1997. Characterisation of integrons and antibiotic resistance genes in Danish multiresistant Salmonella enterica Typhimurium DT104. FEMS Microbiol. Lett. 157:177-181. [DOI] [PubMed] [Google Scholar]
- 36.Schmieger, H. 1999. Molecular survey of the Salmonella phage typing system of Anderson. J. Bacteriol. 181:1630-1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Skuce, R. A., T. P. McCorry, J. F. McCarroll, S. M. Roring, A. N. Scott, D. Brittain, S. L. Hughes, R. G. Hewinson, and S. D. Neill. 2002. Discrimination of Mycobacterium tuberculosis complex bacteria using novel VNTR-PCR targets. Microbiology 148:519-528. [DOI] [PubMed] [Google Scholar]
- 38.Smith, K. L., V. DeVos, H. Bryden, L. B. Price, M. E. Hugh-Jones, and P. Keim. 2000. Bacillus anthracis diversity in Kruger National Park. J. Clin. Microbiol. 38:3780-3784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stanley, T. L., C. D. Ellermeier, and J. M. Slauch. 2000. Tissue-specific gene expression identifies a gene in the lysogenic phage Gifsy-1 that affects Salmonella enterica serovar typhimurium survival in Peyer's patches. J. Bacteriol. 182:4406-4413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Supply, P., S. Lesjean, E. Savine, K. Kremer, D. van Soolingen, and C. Locht. 2001. Automated high-throughput genotyping for study of global epidemiology of Mycobacterium tuberculosis based on mycobacterial interspersed repetitive units. J. Clin. Microbiol. 39:3563-3571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Supply, P., E. Mazars, S. Lesjean, V. Vincent, B. Gicquel, and C. Locht. 2000. Variable human minisatellite-like regions in the Mycobacterium tuberculosis genome. Mol. Microbiol. 36:762-771. [DOI] [PubMed] [Google Scholar]
- 42.Threlfall, E. J. 2000. Epidemic salmonella typhimurium DT 104—a truly international multiresistant clone. J. Antimicrob. Chemother. 46:7-10. [DOI] [PubMed] [Google Scholar]
- 43.van Belkum, A., W. J. Melchers, C. IJsseldijk, L. Nohlmans, H. Verbrugh, and J. F. Meis. 1997. Outbreak of amoxicillin-resistant Haemophilus influenzae type b: variable number of tandem repeats as novel molecular markers. J. Clin. Microbiol. 35:1517-1520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.van Belkum, A., S. Scherer, W. van Leeuwen, D. Willemse, L. van Alphen, and H. Verbrugh. 1997. Variable number of tandem repeats in clinical strains of Haemophilus influenzae. Infect. Immun. 65:5017-5027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.van der Wolf, P. J., T. J. Vercammen, J. J. Geene, A. C. van Exsel, N. H. Peperkamp, M. T. Voets, and A. A. Zeeuwen. 2001. Salmonella typhimurium DT104 septicaemia with meningitis in neonatal piglets. Vet. Q. 23:199-201. [DOI] [PubMed] [Google Scholar]
- 46.Vos, P., R. Hogers, M. Bleeker, M. Reijans, T. van de Lee, M. Hornes, A. Frijters, J. Pot, J. Peleman, M. Kuiper, and M. Zabeau. 1995. AFLP: a new technique for DNA fingerprinting. Nucleic Acids Res. 23:4407-4414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wall, P. G., D. Morgan, K. Lamden, M. Ryan, M. Griffin, E. J. Threlfall, L. R. Ward, and B. Rowe. 1994. A case control study of infection with an epidemic strain of multiresistant Salmonella typhimurium DT104 in England and Wales. Commun. Dis. Rep. Rev. 4:R130-R135. [PubMed]
- 48.Weese, J. S., J. D. Baird, C. Poppe, and M. Archambault. 2001. Emergence of Salmonella typhimurium definitive type 104 (DT104) as an important cause of salmonellosis in horses in Ontario. Can. Vet. J. 42:788-792. [PMC free article] [PubMed] [Google Scholar]
- 49.Weiser, J. N., D. J. Maskell, P. D. Butler, A. A. Lindberg, and E. R. Moxon. 1990. Characterization of repetitive sequences controlling phase variation of Haemophilus influenzae lipopolysaccharide. J. Bacteriol. 172:3304-3309. [DOI] [PMC free article] [PubMed] [Google Scholar]