Abstract
Understanding the complex mechanisms regulating gene expression at the transcriptional and post-transcriptional levels is one of the greatest challenges of the post-genomic era. The MoD (MOtif Discovery) Tools web server comprises a set of tools for the discovery of novel conserved sequence and structure motifs in nucleotide sequences, motifs that in turn are good candidates for regulatory activity. The server includes the following programs: Weeder, for the discovery of conserved transcription factor binding sites (TFBSs) in nucleotide sequences from co-regulated genes; WeederH, for the discovery of conserved TFBSs and distal regulatory modules in sequences from homologous genes; RNAProfile, for the discovery of conserved secondary structure motifs in unaligned RNA sequences whose secondary structure is not known. In this way, a given gene can be compared with other co-regulated genes or with its homologs, or its mRNA can be analyzed for conserved motifs regulating its post-transcriptional fate. The web server thus provides researchers with different strategies and methods to investigate the regulation of gene expression, at both the transcriptional and post-transcriptional levels. Available at http://www.pesolelab.it/modtools/ and http://www.beacon.unimi.it/modtools/.
INTRODUCTION
Understanding the complex mechanisms regulating gene expression is one of the greatest challenges of modern molecular biology. In eukaryotes, gene expression is finely regulated and modulated, at both the transcriptional and post-transcriptional levels. Transcription is regulated by the interaction of transcription factors (TFs) with their corresponding binding sites (TFBSs) (1), mostly located near the transcription start site (TSS) of the gene (i.e. proximal promoter region) or far apart (i.e. enhancers, silencers, etc.). Moreover, recent research has discovered a number of different functions for non-coding RNA molecules, highlighting the need for suitable algorithms and tools for their analysis and comparison aimed at the detection of motifs that are conserved both in structure and (more loosely) in sequence (2). Examples of these functional motifs are the secondary structure signals present in the untranslated regions (UTRs) of mRNAs, often involved in the regulation of their translation and fate (3).
One possible bioinformatics approach to the problem is to select a set of co-regulated genes and look for conserved motifs appearing in their promoters, which are likely to represent binding sites for the common TF(s) regulating them (4). Alternatively, a single gene can be compared with its homologs in other species (or, sometimes, its paralogs) by looking for sequence conservation in the non-coding regions flanking it or within introns (5): regions preserved by evolution are likely to play some role in its regulation. Finally, a set of transcripts, either from genes likely to be subject to the same post-transcriptional regulatory mechanism or from homologous genes, can be examined for conserved motifs, again likely to be functionally active. In this case, however, structure conservation must be taken into account (2).
The MOtif Discovery (MoD) Tools web server provides access to software tools for the discovery of novel motifs that cover these aspects and approaches. It includes Weeder (6,7), for the discovery of conserved motifs in sequences from co-regulated genes; WeederH, for the discovery of motifs and distal motif modules in sequences from homologous genes; and RNAprofile (8), for finding motifs conserved in both sequence and secondary structure in non-coding RNAs.
METHODS
The main page of the web server (Supplementary Data) provides a brief description of the tools implemented. The frames on the left-hand side of the page provide links to the input forms for the online programs (‘Online tools’), to programs for further analysis of the results obtained (‘Additional tools’), and to pages from which to download standalone versions of the tools (‘Downloads’).
WEEDER
Weeder is a software tool for the discovery of conserved TFBSs in nucleotide sequences from co-regulated genes (6,7). A recent comparative assessment of the performance of tools for this task has shown a quite satisfactory performance for Weeder (9). The results of the assessment, however, also highlighted the fact that the different approaches seem to be somewhat complementary: thus, more complete results can be obtained by submitting the same sequences also to methods based on completely different principles from those of Weeder, such as MEME or the Gibbs Sampler [for references and pointers to other tools see (9)]. With respect to the original Weeder web implementation, the new interface allows users to compute an estimate for the P-value associated with a Weeder motif, thus giving a better idea of its significance, and also to perform additional post-processing on their sequence sets by using the motif locator tool.
User Input
The analysis that can be performed by Weeder requires a set of at least two promoters (or UTRs, or in general non-coding regions) from co-regulated genes: the conserved motifs detected can in turn represent instances of TFBSs recognized by the TF(s) regulating the expression of the genes. The size of the sequences should not exceed 1000–1500 bp; moreover, since the annotation of a reliable TSS for a gene is often difficult, the 5′-UTRs (or the full first non-coding exon) should be included. Genes annotated only starting from the ATG codon should be avoided, since in this case the true promoter could be located thousands of base pairs away. Further details on the algorithm are available in the online Supplementary Data.
The input form requires users to input their Email address, two or more sequences in FASTA format and a few intuitive parameters. The web interface first sends a confirmation Email to the address provided and then automatically starts a series of runs of the Weeder algorithm, looking for motifs of length 6 and 8 (if launched in quick mode) or of even length from 6 up to 12 (in normal mode and thorough mode). Upon the completion of the algorithm, results are sent by Email with a link to a dedicated results web page with a friendly graphic layout.
The Output
The highest-scoring motifs of each length considered are shown at the top of the result file. The list of highest-scoring motifs is further processed by the program, as explained in the Supplementary Data (which also shows an example of the output layout), by looking for the most ‘interesting’ ones. The highest-scoring motif of each run, together with those deemed to be ‘interesting’ is reported in detail, under the heading ‘My Advice’. For each, the output shows a frequency matrix built by aligning all the instances of the motif found, as well as a list of its best instances collected from the input sequences using the frequency matrix itself. Another frequency matrix, obtained by aligning only the best occurrences of the motif, is also provided.
Locating motifs in sequences (The ‘Motif Locator’)
The purpose of this program is to locate instances of a given motif in a set of sequences: users have to input a file with the sequences, the motif (e.g. output by Weeder, or any motif describing the consensus of known TFBSs), the maximum number of substitutions allowed, the score percentage threshold with respect to the frequency matrix built (Supplementary Data), and finally whether the search has to be performed on a single strand or on a double strand. Results are output in the same way as for Weeder.
Computing motif P-values (the ‘Motif P-Value Calculator’)
The P-value interface computes a tentative P-value for a Weeder motif according to its score. Users have to input the motif, its Weeder score and statistics on the input sequences that were used to detect the motif (number, size, parameters used with Weeder). If the sequences are of different lengths, the average should be used. The program computes the score that would have been obtained for the same motif and with the same input parameters on a completely random set of sequences of the same size, simply by picking at random the same number of sequences from the promoters of the species selected. This step is performed n (>1000) times, obtaining the number t of ‘random’ runs where the motif had a score higher than the one input. The P-value reported by the interface is defined as P = t/n.
WEEDERH
Another approach to the analysis of gene expression is to compare the regions flanking a given gene, potentially involved in its regulation, with those derived from orthologous genes in other species. This method, also known as phylogenetic footprinting (10), aims at identifying those non-coding regions conserved by evolution, which are likely to play some regulatory function.
From a computational point of view, the problem becomes, rather than one of detecting very subtle similarities, as in the previous case, one of finding conserved motifs or regions in the presence of a much higher level of conservation, deriving from the common evolutionary origin of the sequences investigated. Motif discovery tools such as Weeder can also be applied to this case, but the result is often (e.g. in a human–rodents comparison) a long list of ‘significantly conserved’ motifs that covers virtually all the input.
The idea is that first of all we can expect TFBSs conserved in sequence to be conserved also in their position relative to the gene they regulate: motifs near the TSS of a gene should be near the TSS also in its homologs; similarly, motifs located several thousand basepairs from a gene should also be several thousand basepairs away in the homologs. Moreover, defining an absolute measure of significance for conservation is often impossible since sequence conservation varies greatly according to the species considered, and also according to the genes examined. In WeederH, conservation is measured not in an absolute way but relatively, according to the average degree of conservation of the sequences analyzed, i.e. the average motif score. The idea is that functional elements should be more conserved than the remainder of the sequences. Adding position conservation to the scoring scheme has the effect of permitting the analysis of sequences much longer than the usual promoter size (in our experiments, we obtained successful results for regions of 25–30 kb), looking also for conserved regions far away from the gene that are likely to constitute distal regulatory elements such as enhancers.
The algorithm works on a ‘reference’ sequence, comparing it with one or more homologous sequences, which can come either from orthologous genes in other species or from paralogous genes in the same genome. Motifs in the reference sequence are located by the program. If the size of the reference sequence exceeds 1000 bp the algorithm performs an additional step, trying to identify regions that contain a large number of high-scoring motifs. Since enhancers and cis-regulatory modules in general are usually composed of clusters of TFBSs, a region containing many conserved motifs is in turn a good candidate to represent a conserved enhancer. Further details are available in the online Supplementary Data.
User Input
Users have to input the sequences in separate boxes, together with their organism of origin. The choice between ‘Upstream’ and ‘Downstream’ simply influences the output format. Since in this case the statistical evaluation also considers the position of motifs in the sequences, special care should be taken over the selection of the input sequences. The algorithm, in fact, assumes that all the sequences were derived with respect to the same reference point: thus sequences must be upstream of the TSS of homologous genes, or of the ATG codon, since identifying truly homologous promoters is often very difficult, as shown in (5). The choice of the start codon is, however, also advisable in the case of carefully annotated TSSs, since in many cases multiple TSSs can be annotated for each gene. When genes present a first non-coding exon, this should also be included in the input. Finally, repeats in the input sequences should not be masked.
Program Output
If the submission is successful, the interface shows a confirmation page and, upon completion of the run, results are sent by e-mail. The e-mail contains four links. The first one points to the raw output of the program, containing the ranked list of highest-scoring motifs, with their relative position in the reference sequence and their corresponding regions in each of the homologs. Output motifs do not overlap; i.e. those that would overlap a motif with higher score are omitted. Positions are relative to the end of the sequences (negative) in the case of upstream sequences, relative to the start (positive) otherwise. As explained in the Supplementary Data (with an example of the output), the score associated with each motif is relative, computed according to the average score of the motifs found in the sequences. If the reference input sequence is >1000 bp, at the end of the list of motifs the output contains an additional list of 500 bp regions containing high-scoring motifs, together with their average score.
Another link in the results Email allows users to retrieve the ‘masked’ reference sequence, where all the nucleotides not belonging to a motif are masked with an ‘N’. The masked sequence where only the most significantly conserved motifs are left can be used for further analysis with different tools: e.g. it can be compared with other sequences from co-regulated genes using Weeder.
Using the UCSC genome browser to display results
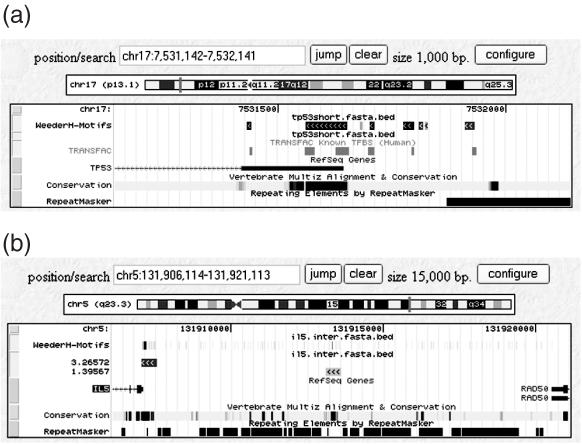
The results e-mail also contains a link that permits results to be displayed within a window of the UCSC genome browser (11), by accessing a page where the user has to input the genomic coordinates of the reference sequence. Input and output files are uploaded automatically. The result is a link to an UCSC genome browser page showing the genomic region corresponding to the reference sequence annotated with motifs and regions selected by WeederH. Moreover, a second track is shown in the browser, corresponding to the 500 bp regions selected by the algorithm, together with their average motif score. Scores are scaled from 0 to 1000 to fit the coloring scheme of the browser: darker regions correspond to motifs or regions with higher score. The tracks (in BED format) can also be downloaded separately. Examples of tracks displaying the output of WeederH are given in Figure 1.
Figure 1.
The output of WeederH, shown within the UCSC genome browser. (a) For the 500 bp upstream and first non-coding exon of the p53 gene of human, mouse and rat. Known TFBSs annotated for the human gene in the TRANSFAC database (14) are also shown. Motifs that do not match the TRANSFAC annotations correspond to the human homologs of sites annotated in the mouse promoter. (b) Obtained from the analysis of the intergenic region upstream of the interleukin-5 gene of human, mouse and rat. The second track from the top shows the 500 bp regions selected by WeederH as containing motifs with significantly high score. The regions selected cover the promoter of the gene and an experimentally validated enhancer at −6500 bp from the gene (15).
RNAPROFILE
RNAProfile takes as input a set of unaligned RNA sequences and outputs the most conserved motifs in the sequences, taking into account both the sequence of the regions and the secondary structures they can form according to thermodynamic rules. We refer the reader to (8) and to the Supplementary Data for further details on the algorithm.
User Input
The basic input form requires users simply to input their sequences and select whether the motif expected should form one or two hairpins. All the parameters for the motifs and those used by the algorithm can be modified in the extended input form. Further details can be obtained online by clicking on each input item on the extended input web page. Also, the extended input form should be used if motifs are expected to appear in a limited subset of the input sequences.
During computation, a temporary page is shown, with a link to the page where the results will be displayed once the computation has ended. The page is automatically refreshed, and results will appear within it as soon as they are ready. Users can, however, save or bookmark the address of the results page for later use.
Program Output
The output shows a list of the 10 highest scoring motifs, each one represented with a sequence logo [built using a tool devised for RNA sequences (12)]. Under the logo, the list of regions that have been used to build the motif is displayed, as well as their secondary structure and corresponding folding energy (E). The ‘fitness’ value (Supplementary Data) is intended to provide an estimate of how well the region fits the motif description: a positive or close-to-zero (even if negative) fitness value means that the region is definitely an instance of the motif described by the profile. Negative values, in contrast, are suspicious. The more negative the value, the more unlikely a region is to be a real instance of the motif. Clicking each region causes the browser to show a graphic representation of its structure. An example is given in Figure 2, showing the iron responsive element [IRE (13)] obtained from the analysis of the 5′-UTR of five ferritin genes. Notice the low fitness score associated with the bottom region, which comes from a plant mRNA lacking the IRE. This region can thus be reasonably regarded as not being a valid motif instance.
Figure 2.
Output of RNAprofile, showing instances of a motif corresponding to the IRE (13), obtained from the analysis of five ferritin mRNA 5′-UTRs. Notice that the last region, with a very low fitness score, is very unlikely to be a true instance of the motif.
CONCLUSIONS
The wealth of genomic data available, complemented by other sources of information on gene expression, have opened new avenues for research. Identifying and characterizing all the elements involved in the regulation of a gene, at both the transcriptional and post-transcriptional level, however, remains one of the most challenging open problems for computer scientists, bioinformaticians and molecular biologists. The tools included in the MoD Tools web server provide researchers with different viewpoints and strategies for their investigations, allowing them to obtain predictions for several types of elements that might be involved in the regulation of one or more genes.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
Acknowledgments
Funding to pay the Open Access publication charges for this article was provided by the Italian Ministry of Education and Research (MIUR) under the FIRB project ‘Laboratorio Italiano di Bioinformatica’.
Conflict of interest statement. None declared.
REFERENCES
- 1.Levine M., Tjian R. Transcription regulation and animal diversity. Nature. 2003;424:147–151. doi: 10.1038/nature01763. [DOI] [PubMed] [Google Scholar]
- 2.Dandekar T., Hentze M.W. Finding the hairpin in the haystack: searching for RNA motifs. Trends Genet. 1995;11:45–50. doi: 10.1016/s0168-9525(00)88996-9. [DOI] [PubMed] [Google Scholar]
- 3.Mignone F., Gissi C., Liuni S., Pesole G. Untranslated regions of mRNAs. Genome Biol. 2002;3 doi: 10.1186/gb-2002-3-3-reviews0004. REVIEWS0004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pavesi G., Mauri G., Pesole G. In silico representation and discovery of transcription factor binding sites. Brief Bioinform. 2004;5:217–236. doi: 10.1093/bib/5.3.217. [DOI] [PubMed] [Google Scholar]
- 5.Prakash A., Tompa M. Discovery of regulatory elements in vertebrates through comparative genomics. Nat. Biotechnol. 2005;23:1249–1256. doi: 10.1038/nbt1140. [DOI] [PubMed] [Google Scholar]
- 6.Pavesi G., Mauri G., Pesole G. An algorithm for finding signals of unknown length in DNA sequences. Bioinformatics. 2001;17:S207–S214. doi: 10.1093/bioinformatics/17.suppl_1.s207. [DOI] [PubMed] [Google Scholar]
- 7.Pavesi G., Mereghetti P., Mauri G., Pesole G. Weeder web: discovery of transcription factor binding sites in a set of sequences from co-regulated genes. Nucleic Acids Res. 2004;32:W199–W203. doi: 10.1093/nar/gkh465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pavesi G., Mauri G., Stefani M., Pesole G. RNAProfile: an algorithm for finding conserved secondary structure motifs in unaligned RNA sequences. Nucleic Acids Res. 2004;32:3258–3269. doi: 10.1093/nar/gkh650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tompa M., Li N., Bailey T.L., Church G.M., De Moor B., Eskin E., Favorov A.V., Frith M.C., Fu Y., Kent W.J., et al. Assessing computational tools for the discovery of transcription factor binding sites. Nat. Biotechnol. 2005;23:137–144. doi: 10.1038/nbt1053. [DOI] [PubMed] [Google Scholar]
- 10.Bulyk M.L. Computational prediction of transcription-factor binding site locations. Genome Biol. 2003;5:201. doi: 10.1186/gb-2003-5-1-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Karolchik D., Baertsch R., Diekhans M., Furey T.S., Hinrichs A., Lu Y.T., Roskin K.M., Schwartz M., Sugnet C.W., Thomas D.J., et al. The UCSC Genome Browser Database. Nucleic Acids Res. 2003;31:51–54. doi: 10.1093/nar/gkg129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gorodkin J., Heyer L.J., Brunak S., Stormo G.D. Displaying the information contents of structural RNA alignments: the structure logos. Comput. Appl. Biosci. 1997;13:583–586. doi: 10.1093/bioinformatics/13.6.583. [DOI] [PubMed] [Google Scholar]
- 13.Theil E.C. The iron responsive element (IRE) family of mRNA regulators. Regulation of iron transport and uptake compared in animals, plants, and microorganisms. Met. Ions Biol. Syst. 1998;35:403–434. [PubMed] [Google Scholar]
- 14.Matys V., Fricke E., Geffers R., Gossling E., Haubrock M., Hehl R., Hornischer K., Karas D., Kel A.E., Kel-Margoulis O.V., et al. TRANSFAC: transcriptional regulation, from patterns to profiles. Nucleic Acids Res. 2003;31:374–378. doi: 10.1093/nar/gkg108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Urwin D.L., Schwenger G.T., Groth D.M., Sanderson C.J. Distal regulatory elements play an important role in regulation of the human IL-5 gene. Eur. J. Immunol. 2004;34:3633–3643. doi: 10.1002/eji.200425279. [DOI] [PubMed] [Google Scholar]