Abstract
Calnuc (nucleobindin) was previously shown to be present both in the cytosol and in the Golgi and to be the major Golgi Ca2+ binding protein. In this study we verified the existence of the cytosolic pool of calnuc and investigated its interaction with Gαi3. Cytosolic calnuc was released by mild digitonin permeabilization. In pulse–chase experiments, the two pools of calnuc had different mobilities, suggesting different posttranslational modifications. That calnuc interacts with Gαi3 in vivo was verified by the finding that Gαi3 could be crosslinked intracellularly to calnuc and co-immunoprecipitated from NIH 3T3 cells stably overexpressing either activated (Q204L) or inactivated (G203A) Gαi3. Binding was Ca2+ and Mg2+-dependent. Calnuc and Gαi3-GFP codistributed primarily in the Golgi region. By yeast two-hybrid analysis, the binding site on Gαi3 for calnuc was mapped to the C-terminal region because removal of the last 12 amino acids (but not 11) abolished the interaction. Peptide competition indicated that calnuc, with its coiled-coil domain constituted by the two EF-hands, binds to Gαi3's C-terminal α5-helix. These results demonstrate that calnuc may play an important role in G protein- and Ca2+-regulated signal transduction events.
Keywords: heterotrimeric G proteins, nucleobindin, signal sequence
Previously we demonstrated that calnuc (nucleobindin) (1, 2), an EF-hand Ca2+ binding protein, is the major Ca2+-binding protein in the Golgi and plays an important role in establishing an agonist-releasable Ca2+ store in the Golgi lumen (3, 4). Two pools of calnuc were found—one associated with the luminal surface of Golgi membranes and the other in the cytosol (3). Thus, calnuc resembles several other signal sequence-containing proteins encoded by a single gene that show dual localizations (5, 6), both in the lumen of cell compartments and in the cytosol (7, 8).
Calnuc was found to interact with several different subfamilies of Gα (3), including Gαi (3, 9) and Gαs (3), using the yeast two-hybrid system. We further pinpointed the Gαi3 binding site on calnuc as the region containing both EF-hands (α helix-loop-α helix) (3), which constitutes an ideal coiled-coil domain (10). Where calnuc binds to Gαi3 remains unknown. A number of functional domains have been identified in Gα. Among them, the C-terminal region is a key determinant of the fidelity of receptor interaction (11, 12) and effector binding (13), as well as constituting a GDP binding pocket (14–16).
In this article we carried out studies designed to further characterize the cytosolic pool of calnuc, which we assume is the pool that interacts with membrane-associated G proteins, and to map the calnuc binding region on Gαi3. Calnuc was found to specifically bind to the C-terminal α5 helix region of Gαi3, and the interaction is regulated by Ca2+ and Mg2+.
Materials and Methods
Materials.
Polyclonal antibody (F-5059) against recombinant, full length calnuc was generated and affinity purified as described (3). Affinity-purified rabbit IgG against Gαi3 (EC) and NIH 3T3 fibroblasts stably overexpressing activated (Q204L) or inactivated (G203A) Gαi3 (17) were generous gifts from A. Spiegel (National Institute of Diabetes and Digestive and Kidney Diseases) and S. Hermouet (Hopitaux De Nantes, Nantes, France). Green fluorescent protein (GFP) cDNA was kindly provided by Roger Tsien (University of California, San Diego). Cross-absorbed Texas Red-conjugated donkey anti-rabbit F(ab′)2 was obtained from Jackson ImmunoResearch, and affinity purified goat anti-rabbit IgG (H+L) conjugated to horseradish peroxidase was from Bio-Rad. HPLC purified synthetic peptides were obtained from Research Genetics (Huntsville, AL). All chemicals were from Sigma except as indicated.
Digitonin Permeabilization and Assessment of Lactate Dehydrogenase (LDH) Activity.
Normal rat kidney (NRK) cells were plated on 96-well plates (1 × 104 cells/well) and were cultured overnight at 37°C as described (3). After removal of the culture medium, cells were permeabilized by addition of 200 μl of digitonin solution in Tris-buffered saline (20–50 μg/ml digitonin/150 mM NaCl/20 mM Tris⋅HCl, pH. 7.4) for 5 min at 4°C (18). The amount of LDH released after digitonin permeabilization was determined by using a Cytotoxicity Detection Kit (Roche Molecular Biochemicals) and a Vmax Kinetic Microplate Reader (λ = 490 nm) (Molecular Devices). The amount of cytosolic calnuc released was similarly assessed on cells cultured on 100-mm dishes to which 1.5 ml of digitonin solution was added for 5 min at 4°C. The digitonin solution was collected and centrifuged (100,000 × g/1 h), and the released proteins were precipitated with 10% trichloroacetic acid. The trichloroacetic acid precipitate was resolved by SDS/PAGE and was immunoblotted with affinity-purified anti-calnuc IgG as described (3).
Pulse–Chase and Immunoprecipitation.
NRK cells (1 × 107 cells/100-mm dish) were preincubated in 10 ml Met- and Cys-free DMEM (ICN) for 8 min at 37°C, followed by pulse-labeling for 5 min in 2 ml Tris-buffered saline containing 150 μCi/ml l-[35S]Met (1,000 Ci/mM; DuPont/NEN). Cells were then rinsed with DMEM containing unlabeled methionine (3 mg/ml), followed by chase in the same media (5–30 min). Cytosolic (100,000 × g supernatant) and membrane (100,000 × g pellet) fractions were prepared from postnuclear supernatants as described (3, 4), using one dish (1 × 107 cells) for each time point. Fractions were solubilized in RIPA buffer [0.1% SDS, 0.5% Triton X-100, 0.5% sodium deoxycholate in Tris-buffered saline (pH 7.4)], followed by immunoprecipitation of the lysates with 5 μg anti-calnuc IgG and 10 μl Protein A-Sepharose CL-4B (Amersham Pharmacia) in RIPA buffer. Beads were washed (four times, 5 min each) at 4°C with RIPA buffer, and the immune complexes were separated by SDS/PAGE and were exposed to Kodak Biomax MR film at −80° C for 3 days for autoradiography.
Co-Immunoprecipitation of Calnuc and Gαi3.
Four dishes of NIH 3T3 cells (1 × 107 cells/100-mm dish) stably overexpressing activated (Q204L) or inactivated (G203A) Gαi3 were lysed in 300 μl co-immunoprecipitation (coIP) buffer (0.5% Triton X-100/25 mM Hepes, pH 7.4) containing either 2 mM Ca2+/15 mM Mg2+ (19) or 5 mM EDTA in the presence of protease inhibitors (3) for 30 min at 4°C. The lysate was supplemented to 0.1 M NaCl, and 1 M Non-Detergent Sulfobetaine (NDSB-195) (Calbiochem) was added to increase soluble protein recovery (20). After centrifugation (16,000 × g for 15 min), the lysates were precleared with preimmune serum coupled to Protein A-Sepharose CL-4B and subsequently were incubated with 5 μg of affinity-purified anti-calnuc IgG followed by 10 μl Protein A beads, each for 1 h at 4°C. Beads were washed four times (5 min each) at 4°C with coIP buffer containing 1 M NDSB-195 and 0.1 M NaCl. Eluted proteins were separated by SDS/10% PAGE under nonreducing conditions, followed by immunoblotting with affinity-purified anti-Gαi3 IgG (EC) and enhanced chemiluminescence (ECL) analysis (Amersham Pharmacia).
Crosslinking of Calnuc and Gαi3 in Intact Cells.
To maintain the intracellular Mg2+/Ca2+ concentration at a constant level, four 100-mm dishes of confluent NIH 3T3 cells stably overexpressing activated or inactivated Gαi3 were incubated with 10 μM ionomycin containing either 2 mM Ca2+/15 mM Mg2+ or 5 mM EDTA (21, 22) in 2.7 ml reaction buffer containing 150 mM NaCl/20 mM Hepes (pH 7.4) at 37°C for 15 min. The membrane-permeant crosslinker, dithiobis (succinimidylpropionate) (Pierce), was then added to a final concentration of 2 mM for 30 min at 37°C. The reaction was quenched with 3 ml of 100 mM Tris⋅HCl (pH 7.5) for 20 min at 37°C. Cells were lysed with 1% Triton X-100 in Tris-buffered saline containing 1 M NDSB-195 at 4°C for 30 min in the presence of either 2 mM Ca2+/15 mM Mg2+ or 5 mM EDTA. Precleared lysates were immunoprecipitated with anti-calnuc IgG and Protein A beads, as for the coIP experiments, followed by SDS/PAGE and immunoblotting with anti-Gαi3 IgG.
Transient Overexpression of Gαi3-GFP in EcR-CHO-Calnuc Cells.
Gαi3-GFP cDNA was prepared by ligation of GFP cDNA to the 3′ end of Gαi3 cDNA and was subcloned into the pIND vector (Invitrogen) at EcoRI/NotI restriction sites with HindIII as an internal linker. EcR-CHO-Calnuc cells (4) were transiently transfected with Gαi3-GFP cDNA using lipofectamine (GIBCO/BRL) as described (4). Overexpression of calnuc and Gαi3-GFP (transient) was induced with 10 μM ponasterone A (Invitrogen) for 48 h, followed by treatment or not with ionomycin/Ca2+/Mg2+ as described above. Cells were subsequently processed for immunofluorescence localization of calnuc (4).
In Vivo Interactions in the Yeast Two-Hybrid System.
Wild-type and mutant Gαi3 cDNAs were generated by PCR with appropriate primers (sequences available on request) and were subcloned into the pAS2–1 DNA-BD (bait) vector (CLONTECH) at EcoRI/SmaI restriction sites. Gαi3-GFP and Gαi3(ΔC12)-GFP cDNAs were subcloned into pAS2–1 at EcoRI/PstI sites with HindIII as an internal linker. Calnuc/pACT2 AD (prey) was constructed as described (3). All constructs were verified by automated DNA sequencing (Center for AIDS Research, University of California, San Diego). For one-to-one interaction, Gαi3/pAS2–1(bait), calnuc/pACT2 (prey), and carrier DNA were co-transformed into competent yeast cells (strain SFY 526) (CLONTECH). Four days after transformation, the colony-lift assay was performed using β-galactosidase and 5-bromo-4-chloro-3-indolyl-β-d-galactoside.
Preparation and Purification of His6-Calnuc and His6-Gαi3.
His6-calnuc (4) and His6-Gαi3 (23) were prepared and purified as described earlier except that His6-calnuc protein (eluted with 100 mM imidazole) was dialyzed and stored in 100 mM NaCl/20 mM Hepes (pH 7.4).
Peptide Competition Assays.
[35S]Gαi3 was obtained by in vitro translation using a TNT T7 Quick Coupled Transcription/Translation System (Promega) and l-[35S]Met (10 mCi/ml) (Amersham Pharmacia, cell labeling grade, 50 μCi/reaction). Rat Gαi3 cDNA subloned into the pcDNA-3 vector was used as a template (24). Purified His6-calnuc was covalently conjugated to cyanogen bromide-activated Sepharose 4B (Pharmacia) as described (3) to a final concentration of 2 μg of protein/1 μl of beads. [35S]Gαi3 (3 μl, 15,000 cpm) were incubated with 10 μl His6-calnuc-Sepharose 4B in the presence and absence of 2 mM of each of four HPLC-purified synthetic peptides or 160 μg His6-Gαi3 in a total volume of 250 μl pull-down buffer (0.5% Triton X-100/100 mM NaCl/20 mM Hepes, pH 7.4) containing either 2 mM Ca2+/15 mM Mg2+ or 5 mM EDTA. Incubation was carried out at 4°C for 1 h, and the beads were subsequently washed (four times over 20 min) with pull-down buffer, followed by SDS/PAGE and autoradiography.
Results
Existence of Cytosolic and Membrane-Associated Pools of Calnuc.
We previously demonstrated, based on immunoblotting of cell fractions, that there are two pools of calnuc, one cytosolic and the other membrane-associated, found in the Golgi lumen (3, 4). To provide further confirmatory evidence for the existence of a cytosolic pool of calnuc, we permeabilized NRK cells with low concentrations of digitonin and analyzed the composition of the released cytosol under conditions known to permeabilize the plasma membrane but to maintain Golgi membranes intact (18, 25), thus avoiding release of the Golgi pool of calnuc. Fig. 1A shows that there was a linear increase in the amount of the cytosolic marker LDH released with increasing concentrations of digitonin (20–50 μg/ml). Under the same conditions, increasing amounts of calnuc were also released (Fig. 1B), indicating that, in addition to its association with the luminal side of Golgi membranes, calnuc is also found in the cytosol (3).
Figure 1.
Validation of the existence of cytosolic and membrane-associated pools of calnuc. (A) Release of LDH from permeabilized cells. NRK cells were permeabilized with the indicated concentrations of digitonin, and supernatant was collected for assessment of LDH activity. LDH release increases linearly with increasing concentrations of digitonin. Results (mean ± SD) represent the average of values obtained in three separate experiments performed in triplicate. (B) Release of cytosolic calnuc after digitonin treatment. NRK cells were permeabilized with digitonin as in A. Released calnuc was precipitated from the supernatant with 10% trichloroacetic acid, separated by SDS/PAGE, and immunoblotted with affinity-purified anti-calnuc IgG. Increasing amounts of calnuc are released with increasing digitonin concentrations. (C) Pulse–chase experiments. NRK cells were pulse-labeled with [35S]Met (300 μCi/dish) for 5 min and chased for the indicated time intervals. [35S]calnuc was immunoprecipitated from cytosolic (S) and membrane (P) fractions, followed by autoradiographic analysis. The membrane-associated pool and the cytosolic pool have the same apparent mobility at the beginning of the chase (0), whereas after 5 and 10 min of chase, the membrane pool runs faster than the cytosolic pool. After 30 min of chase, the membrane pool again has approximately the same mobility as the cytosolic pool. Experiments were repeated three times.
To determine the relationship between the two pools of calnuc, we carried out pulse–chase experiments (Fig. 1C). When immunoprecipitation was carried out on membrane and cytosolic fractions from NRK cells metabolically labeled with [35S]Met, calnuc precipitated from the membrane and cytosolic fractions showed the same mobility by SDS/PAGE at the end of the pulse. However, after 5 and 10 min of chase, the membrane pool of calnuc ran faster than the cytosolic pool. After 30 min of chase, the membrane pool again showed approximately the same mobility as that of cytosolic calnuc. These findings indicate that the cytosolic and membrane pools of calnuc show different mobilities by SDS/PAGE at early time points after synthesis. This indicates that the two pools are different and suggests that they undergo distinct posttranslational modifications.
Calnuc Specifically Interacts with Gαi3, and the Interaction Is Ca2+/Mg2+-Dependent.
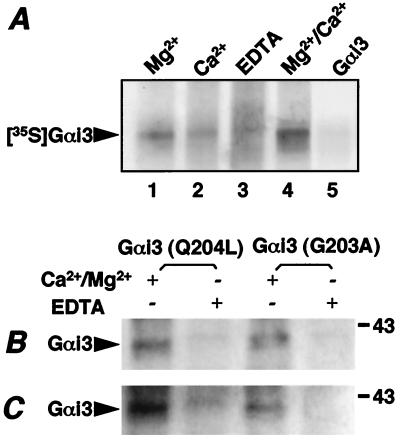
We next examined whether calnuc interacts with Gαi3 by carrying out in vitro pull down assays with calnuc-Sepharose 4B in the presence or absence of divalent cations. Fig. 2A shows that [35S]Gαi3 bound to calnuc in the presence of 15 mM Mg2+ (lane 1), 2 mM Ca2+ (lane 2), or both (lane 4). The binding was strongest in the presence of both Mg2+ and Ca2+ and was weakest in the presence of Ca2+ alone. The interaction between calnuc and [35S]Gαi3 was inhibited by 5 mM EDTA (Fig. 2A, lane 3). It was also inhibited by excess unlabeled recombinant Gαi3 (Fig. 2A, lane 5) in the presence of Ca2+/Mg2+.
Figure 2.
Calnuc specifically interacts with both activated and inactivated Gαi3 in vivo, and the interaction is Ca2+/Mg2+-dependent. (A) Pull-down assays. In vitro translated [35S]Gαi3 was incubated with calnuc-Sepharose 4B at 4°C for 1 h in the presence of 15 mM Mg2+ (lane 1), 2 mM Ca2+ (lane 2), 15 mM Mg2+ + 2 mM Ca2+ (lanes 4 and 5), or 5 mM EDTA (lane 3), followed by extensive washing, SDS/PAGE separation, and autoradiographic analysis. Calnuc interacts with [35S]Gαi3 in the presence of Mg2+, Ca2+, or both, but weak interaction is observed in the presence of Ca2+ alone. Binding is abolished by 5 mM EDTA as well as excess unlabeled Gαi3 (lane 5). (B) coIP of Gαi3 and calnuc. NIH 3T3 cells overexpressing Gαi3(Q204L) (activated) and Gαi3(G203A) (inactivated) were lysed with 0.5% TX-100 at 4°C for 30 min in the presence of 2 mM Ca2+/15 mM Mg2+ or 5 mM EDTA. coIP was performed on the cell lysates with anti-calnuc IgG in the presence of 1 M NDSB-195 and protein A beads, as described in Materials and Methods, followed by immunoblotting with anti-Gαi3 IgG. Calnuc interacts with both activated and inactivated Gαi3 in the presence (+) but not in the absence (−) of 2 mM Ca2+/15 mM Mg2+, suggesting that the interaction between calnuc and Gαi3 is Ca2+/Mg2+-dependent but does not depend on the state of activation of Gαi3. (C) Crosslinking of calnuc and Gαi3. NIH 3T3 cells overexpressing Gαi3(Q204L) and Gαi3(G203A) cells were treated with 10 μM ionomycin plus either 2 mM Ca2+/15 mM Mg2+ or 5 mM EDTA. Crosslinking was carried out by using the membrane permeant crosslinker dithiobis (succinimidylpropionate) (2 mM), followed by lysis with 1% TX-100 and immunoprecipitation with anti-calnuc IgG as above. Similar results to those in B were obtained. Both coIP and crosslinking experiments were repeated four times.
To validate these results in living cells, we carried out immunoprecipitation experiments in mouse 3T3 fibroblasts overexpressing either activated (Q204L) or inactivated (G203A) Gαi3 (17). We found that Gαi3 could be co-immunoprecipitated with calnuc from cells overexpressing either activated or inactivated Gαi3 (Fig. 2B). Again, the interaction was Ca2+- and Mg2+-dependent.
To further confirm that cytosolic calnuc binds to Gαi3 in vivo, we performed crosslinking experiments on intact cells by using the membrane-permeant crosslinker dithiobis (succinimidylpropionate). To maintain the intracellular divalent cation concentration similar to that found to be optimal for coIP, crosslinking was performed in the presence of 10 μM ionomycin and either 2 mM Ca2+/15 mM Mg2+ or 5 mM EDTA (21). Fig. 2C shows that, in the presence of Ca2+/Mg2+, Gαi3 is efficiently cross-linked to calnuc in cells overexpressing either activated (Q204L) or inactivated (G203A) Gαi3. From these results obtained by two different approaches, we conclude that (i) calnuc specifically binds to Gαi3 in vivo, and (ii) the interaction is Ca2+/Mg2+-dependent but does not depend on the state of activation of Gαi3. The latter is in agreement with results previously reported by us (3) and others (9).
Distribution of Calnuc and Gαi3 in EcR-CHO Cells Stably Overexpressing Calnuc.
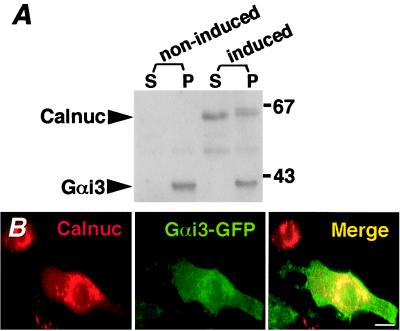
When EcR-CHO-Calnuc cells stably overexpressing calnuc (4) were induced with 10 μM ponasterone A for 48 h, the ratio of the cytosolic to the membrane-associated pool of calnuc was ≈60/40, with cytosolic calnuc showing a faster mobility than that of membrane-associated calnuc (Fig. 3A). Overexpression of calnuc did not significantly affect the distribution of endogenous Gαi3, which was detected only in the membrane fraction in both induced and noninduced cells.
Figure 3.
Localization of Calnuc and Gαi3. (A) Expression of Gαi3 in non-induced and induced (10 μM ponasterone A for 48 h) EcR-CHO-Calnuc cells stably overexpressing calnuc. Cytosolic (100,000 × g supernatant) and membrane (100,000 × g pellet) fractions were prepared and immunoblotted with anti-calnuc IgG and affinity-purified anti-Gαi3 IgG (EC). The ratio of the cytosolic (S) and membrane-associated (P) pools of overexpressed calnuc is ≈60:40. The cytosolic pool shows a faster mobility than that of the membrane pool, which consists of two bands. Overexpression of calnuc does not significantly affect the level and distribution of endogenous Gαi3, which is found exclusively in the membrane fraction (P). However, the mobility of Gαi3 is reduced slightly. (B) Immunofluorescence. EcR-CHO-Calnuc cells were transiently transfected with Gαi3-GFP cDNA, followed by induction with 10 μM ponasterone A for 48 h. To maintain intracellular divalent ion concentration, cells were treated with 10 μM ionomycin/2 mM Ca2+/15 mM Mg2+, followed by fixation in 2% formaldehyde and incubation with anti-calnuc IgG and Texas Red-conjugated donkey anti-rabbit F(ab)′2. Overexpressed calnuc (Left) was detected in the Golgi region and in the cytoplasm. Gαi3-GFP (Middle) was found on the plasma membrane as well as in the Golgi region. The merged images (Right) indicate that the distribution of calnuc and Gαi3 partially overlap mainly in the Golgi region. (Bar = 10 μm.)
To determine whether calnuc and Gαi3 colocalize by immunofluorescence, EcR-CHO-Calnuc cells were transiently transfected with Gαi3-GFP and were induced with ponasterone A (10 μM for 48 h), followed by treatment with ionomycin plus Ca2+/Mg2+. Cells were subsequently fixed and immunostained with anti-calnuc IgG. The majority of the calnuc was found in the Golgi region whereas Gαi3-GFP was found on the plasma membrane as well as in the Golgi region (Fig. 3B). The distribution of Gαi3-GFP and calnuc was similar in the presence or absence of divalent cations. Thus, the distribution of calnuc and Gαi3-GFP partially overlap, primarily in the Golgi region.
Calnuc Interacts with the C-terminal Region of Gαi3.
To determine the calnuc binding region on Gαi3, truncation analysis was performed by using the yeast two-hybrid system. We first examined whether the N-terminal membrane anchoring motifs of Gαi3 including its myristoylation (Gly2) and palmitoylation (Cys3) sites (see Fig. 4) are involved in its interaction with calnuc. Site-directed mutagenesis of the myristoylation site (G2A), the palmitoylation site [C3A(S)], or both [G2A, C3A(S)] did not change the binding of Gαi3 to calnuc (Fig. 5A). Next we carried out step-wise truncation of the N terminus of Gαi3. We found that, after removal of up to 26 amino acids, Gαi327–354 still interacts with calnuc, indicating that the N-terminal region of Gαi3 is not likely to constitute a calnuc binding site.
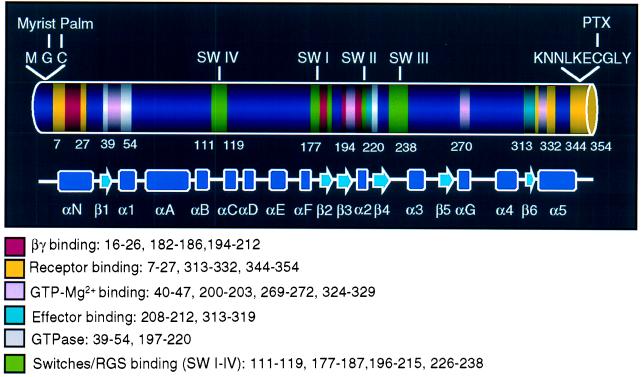
Figure 4.
Primary and secondary structures of Gαi1. The primary and secondary structure as well as functional domains of Gαi1 (12, 28), which are similar to that of Gαi3, are indicated in the diagram. The N terminus (amino acids 1–27) is involved in membrane anchoring (myristoylation and palmitoylation) (35), as well as receptor (11, 12, 26) and βγ (12) binding. The C terminus has been shown to participate in binding to effectors (13) and GDP/GTP (31, 32) as well as to receptors (11, 12, 26). The switch regions (I–III) constitute the binding site for RGS proteins (RGS 4) (41) and adenylyl cyclase (13), as well as βγ subunits (12).
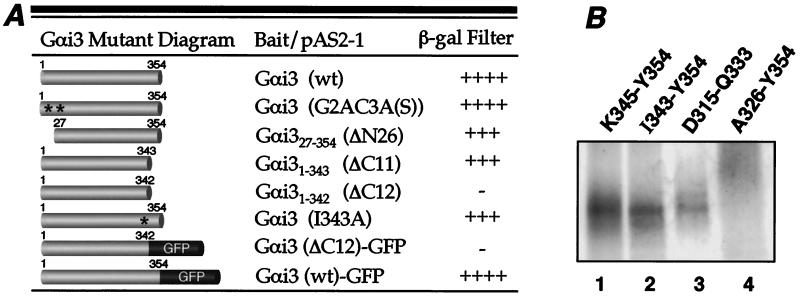
Figure 5.
Calnuc interacts with the C-terminal α5-helix of Gαi3. (A) Two-hybrid assays. Gαi3 mutants were subcloned into the pAS2–1 bait vector, and calnuc was constructed in the pACT2 prey vector. The β-galactosidase filter assay was performed on (Leu−, Trp−) plates, and color was scored after 2 h. (−, negative; +, weak; ++, intermediate; +++, strong; ++++, very strong). Gαi3 mutants in which N-terminal sites for both myristoylation and palmitoylation were mutated [G2AC3A(S)] or the first 26 amino acids were truncated (ΔN26) bind to calnuc as strongly as wild-type (wt) Gαi3. Similarly, truncation of the last 11 C-terminal amino acids (Gαi3 ΔC11) does not reduce Gαi3 binding to calnuc whereas the interaction is completely abolished by deletion of the last 12 C-terminal amino acids (Gαi3 ΔC12). However, Gαi3 (I343A) still strongly binds to calnuc. Addition of GFP to the Gαi3 mutant with the C-terminal 12 amino acids deleted (Gαi3 ΔC12-GFP) does not restore binding to calnuc. Full length Gαi3 (wt)-GFP shows strong interaction with calnuc. Reproducible results were obtained in three separate experiments. (B) Peptide competition assays. 2 mM of each of four synthetic peptides and in vitro translated [35S]Gαi3 were incubated with calnuc-Sepharose 4B in the presence of 15 mM Mg2+/2 mM Ca2+ as described in Fig. 2A, followed by SDS/PAGE and autoradiographic analysis. The peptides K345-Y354 (lane 1), I343-Y354 (lane 2), and D315-Q333 (lane 3) are not able to compete [35S]Gαi3 binding to calnuc whereas peptide A326-Y354 (lane 4) completely inhibits calnuc-Gαi3 interaction.
Next we examined the effects of sequential C-terminal truncation on Gαi3's interaction with calnuc (Fig. 5A). Deletion of 5–11 amino acids had no effect as Gαi31–343 (ΔC11) bound as strongly as full length Gαi3, indicating that the C-terminal receptor binding region (amino acids 345–354) is not the binding site for calnuc. However, deletion of 12 amino acids (Gαi31–342 Δ12) completely abolished binding to calnuc. To rule out that lack of binding is not caused by deletion of the single amino acid (I343), we examined ability of the mutant Gαi3 (I343A) to bind calnuc and found that it interacts with calnuc as strongly as Gαi31–343 (ΔC11). Addition of GFP to the C terminus of Gαi31–342 (Δ12) did not restore its binding capability, although full length Gαi3-GFP strongly interacted with calnuc.
Calnuc Binds to the C-terminal α5-Helix of Gαi3.
Because truncation experiments indicated that the C terminus of Gαi3 is important for binding, we adopted a peptide competition strategy similar to that of Hamm et al. (26) to precisely pinpoint the calnuc binding site. We carried out pull-down assays with in vitro translated [35S]Gαi3 and calnuc covalently coupled to Sepharose 4B in the presence of several overlapping peptides from the C-terminal region of Gαi3. First, we tested the ability of peptides corresponding to the C-terminal receptor binding regions of Gαi3 (Fig. 5A), KNNLKECGLY (K345-Y354) (Fig. 5B, lane 1) and DTKEVYTHFTCATDTKNVQ (D315-Q333) (12) (lane 3), to inhibit the calnuc–Gαi3 interaction. Neither of these peptides inhibited their interaction. Next we examined whether a peptide corresponding to the last 12 amino acids of Gαi3, IIKNNLKECGLY (I343-Y354), inhibits binding. Interestingly, this peptide did not compete Gαi3 binding to calnuc. This result plus the observation obtained from the yeast-two hybrid assay suggest that a region within the α5-helix is involved in calnuc binding. This assumption was verified by the finding that a peptide consisting of the last 28 amino acids (ATDTKNVQFVFDAVTDVIIKNNLKECGLY, A326-Y354) of Gαi3, containing its entire α5-helix of Gαi3 (T329-N347) (12), completely inhibited interaction (Fig. 5B, lane 4).
Discussion
Calnuc was previously demonstrated to interact with Gα subunits, i.e., Gαi (3, 9) and Gαs (3), and was found both in the cytosol and on Golgi membranes facing the Golgi lumen (3). Our previous studies focused on the membrane-associated pool of calnuc, which is the major Ca2+-binding protein in the Golgi and plays an important role in agonist releasable Ca2+ storage in the Golgi (4). In this study, we investigated the cytosolic pool of calnuc and further characterized the calnuc–Gαi3 interaction.
First it was necessary to demonstrate that calnuc in the cytosol was not the result of leakage from the Golgi during homogenization. This was ruled out by results obtained from digitonin permeabilization and pulse–chase experiments. We found that calnuc and LDH, a cytosolic protein marker, are released in parallel after mild permeabilization of the plasma membrane with low concentrations of digitonin, which maintain Golgi and endoplasmic reticulum membranes intact (18, 25). We also showed in pulse–chase experiments that cytosolic and membrane-associated pools of calnuc have different mobilities in the first 30 min after synthesis, demonstrating the distinct nature of the two pools and suggesting that they undergo different posttranslational modifications. These results validate that calnuc is indeed present in the cytosol.
An increasing number of proteins bearing signal sequences targeting newly synthesized protein to the endoplasmic reticulum have been reported to be localized and function in both the cytosol and the secretory pathway. For example, the plasminogen activator inhibitor 2 (PAI-2) (5, 6) undergoes variable translocation, with some of the protein being translocated into the rough endoplasmic reticulum and some remaining in the cytosol. Several potential mechanisms have been proposed to explain this dual localization, including alternative splicing or regulation of the signal sequence (5–8). Because only one size of calnuc mRNA has been identified (27), alternative splicing can be ruled out, and regulation of its signal sequence seems more likely. The protein retained in the cytosol might subsequently undergo additional posttranslational modifications, which might explain the mobility shifts of [35S]calnuc shown in pulse–chase studies (Fig. 1C). The detailed mechanism for generating membrane-associated and cytosolic pools of calnuc remains to be established.
Results obtained from the C-terminal truncation of Gαi3 showed that deletion of the last 12 amino acids abolished its interaction with calnuc. However, deletion of the last 11 amino acids had no effect, indicating that calnuc may bind to another C-terminal segment close to the last 11 amino acids. Alternatively, removal of 12 amino acids may disrupt the structure of the entire Gαi3 subunit (16). To rule out that lack of binding of the mutant with the last 12 amino acids deleted was caused by disruption of the entire Gαi3 subunit and to precisely pinpoint the calnuc binding site in the C-terminal region, a strategy similar to that used previously by Hamm et al. (26) was applied using synthetic peptides. Peptide competition indicated that neither the last 12 amino acids (amino acids 343–354) nor the two C-terminal receptor binding regions (amino acids 315–333 and 345–354) competed Gαi3 binding to calnuc. However, a peptide (amino acids 326–354) including the entire α5 helix (amino acids 329–347) as well as one of the C-terminal receptor binding regions (amino acids 345–354) (12, 28) completely inhibited Gαi3's ability to interact with calnuc. Because the peptide representing the receptor binding region (amino acids 345–354) did not compete Gαi3 binding to calnuc, we believe that the α5 helix (amino acids 329–347) constitutes the calnuc binding site. The C-terminal α5 helix was reported to be stabilized by the interaction between its conserved V342-I343-I344 and V34, L195, A221, and Y321 (29) and/or a salt bridge between the α5 helix (Asp337/Asp341) and β2/β3 loop (Lys192) (30). Therefore, it seems reasonable to speculate that abolishing Gαi3's binding to calnuc by deletion of its C-terminal 12 amino acids may be caused by disruption of the α5 helix. Interestingly, the receptor mimetics, compound 48/80 and mastoparan, have recently been reported to bind and to unwind the α5 helix in Gαi1 (30), indicating that the C-terminal α5 helix may constitute the binding site for several molecules that modulate Gα activity. Because the C-terminal region of Gα is particularly interesting due to its involvement in receptor and effector binding (12, 13, 28) as well as constituting a GDP binding pocket (31, 32), it will be of interest to investigate whether calnuc can regulate Gα or has other functions.
In this study, we demonstrated that binding of calnuc to Gαi3 is Ca2+- and Mg2+-dependent, as they showed a strong interaction in the presence of both Ca2+ and Mg2+, a weak interaction in the presence of Mg2+ or Ca2+ alone, and their interaction was abolished in the presence of EDTA. It was reported previously that calnuc undergoes a conformational change after Ca2+ binding to its EF-hand motifs (33) which constitute an ideal coiled-coil domain (COILS, European Molecular Biology Network-Swiss Node) (10). Mg2+ is also required to maintain Gα activity (31, 34, 35). Therefore, it is reasonable to speculate that Ca2+ and Mg2+ are necessary to maintain the appropriate secondary structures of calnuc and Gαi3 required for their interaction.
Where do calnuc and Gαi3 interact in vivo? Gα subunits are localized on the plasma membrane as well as on intracellular membranes such as Golgi membranes (36–38), as shown here for Gαi3-GFP (see Fig. 3B), where they regulate vesicular trafficking (39, 40). Cytosolic calnuc may interact with Gαi3 either at the plasma membrane or on the Golgi facing the cytosol. It will be of interest to further investigate where the calnuc-Gαi3 interaction occurs and how their interaction is regulated.
In summary, the existence of a cytosolic pool of calnuc and its ability to specifically bind to the C-terminal α5 helix of Gαi3 was demonstrated in this study. The interaction is Ca2+/Mg2+-dependent but independent of the state of activation of Gαi3. Calnuc is the first EF-hand Ca2+ binding protein shown to interact with Gαi3. Further investigation is needed to shed light on the detailed roles of calnuc in G protein and Ca2+-related signal transduction events in cells.
Acknowledgments
T.W. is the recipient of a fellowship from the Deutsche Forschungsgemeinschaft (DFG) (Germany). This work was supported by National Institutes of Health Grants DK17780 and CA58689 to M.G.F.
Abbreviations
- coIP
co-immunoprecipitation
- GFP
green fluorescent protein
- LDH
lactate dehydrogenase
References
- 1.Miura K, Titani K, Kurosawa Y, Kanai Y. Biochem Biophys Res Commun. 1992;187:375–380. doi: 10.1016/s0006-291x(05)81503-7. [DOI] [PubMed] [Google Scholar]
- 2.Wendel M, Sommarin Y, Bergman T, Heinegard D. J Biol Chem. 1995;270:6125–6133. doi: 10.1074/jbc.270.11.6125. [DOI] [PubMed] [Google Scholar]
- 3.Lin P, Le-Niculescu H, Hofmeister R, McCaffery J M, Jin M, Henneman H, McQuistan T, De Vries L, Farquhar M G. J Cell Biol. 1998;141:1515–1527. doi: 10.1083/jcb.141.7.1515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lin P, Yao Y, Hofmeister R, Tsien R Y, Farquhar M G. J Cell Biol. 1999;145:279–289. doi: 10.1083/jcb.145.2.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Danpure C J. Trends Cell Biol. 1995;5:230–238. doi: 10.1016/s0962-8924(00)89016-9. [DOI] [PubMed] [Google Scholar]
- 6.Belin D, Bost S, Vassalli J D, Strub K. EMBO J. 1996;15:468–478. [PMC free article] [PubMed] [Google Scholar]
- 7.Hegde R S, Lingappa V R. Trends Cell Biol. 1999;9:132–137. doi: 10.1016/s0962-8924(99)01504-4. [DOI] [PubMed] [Google Scholar]
- 8.Martoglio B, Dobberstein B. Trends Cell Biol. 1998;8:410–415. doi: 10.1016/s0962-8924(98)01360-9. [DOI] [PubMed] [Google Scholar]
- 9.Mochizuki N, Hibi M, Kanai Y, Insel P A. FEBS Lett. 1995;373:155–158. doi: 10.1016/0014-5793(95)01031-9. [DOI] [PubMed] [Google Scholar]
- 10.McLachlan A D, Karn J. J Mol Biol. 1983;164:605–626. doi: 10.1016/0022-2836(83)90053-0. [DOI] [PubMed] [Google Scholar]
- 11.Bourne H R. Curr Opin Cell Biol. 1997;9:134–142. doi: 10.1016/s0955-0674(97)80054-3. [DOI] [PubMed] [Google Scholar]
- 12.Lambright D G, Sondek J, Bohm A, Skiba N P, Hamm H E, Sigler P B. Nature (London) 1996;379:311–319. doi: 10.1038/379311a0. [DOI] [PubMed] [Google Scholar]
- 13.Sunahara R K, Tesmer J J, Gilman A G, Sprang S R. Science. 1997;278:1943–1947. doi: 10.1126/science.278.5345.1943. [DOI] [PubMed] [Google Scholar]
- 14.Onrust R, Herzmark P, Chi P, Garcia P D, Lichtarge O, Kingsley C, Bourne H R. Science. 1997;275:381–384. doi: 10.1126/science.275.5298.381. [DOI] [PubMed] [Google Scholar]
- 15.Yang C S, Skiba N P, Mazzoni M R, Hamm H E. J Biol Chem. 1999;274:2379–2385. doi: 10.1074/jbc.274.4.2379. [DOI] [PubMed] [Google Scholar]
- 16.Denker B M, Schmidt C J, Neer E J. J Biol Chem. 1992;267:9998–10002. [PubMed] [Google Scholar]
- 17.Hermouet P, De Mazancourt P, Spiegel A M. Cell Signalling. 1993;5:215–225. doi: 10.1016/0898-6568(93)90072-t. [DOI] [PubMed] [Google Scholar]
- 18.Plutner H, Davidson H W, Saraste J, Balch W E. J Cell Biol. 1992;119:1097–1116. doi: 10.1083/jcb.119.5.1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Berman D M, Wilkie T M, Gilman A G. Cell. 1996;86:445–452. doi: 10.1016/s0092-8674(00)80117-8. [DOI] [PubMed] [Google Scholar]
- 20.Vuillard L, Braun-Breton C, Rabilloud T. Biochem J. 1995;305:337–343. doi: 10.1042/bj3050337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Duncan J S, Burgoyne R D. Biochem J. 1996;317:487–493. doi: 10.1042/bj3170487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kolber M A, Haynes D H. Biophys J. 1981;36:369–391. doi: 10.1016/S0006-3495(81)84738-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fischer T, Elenko E, McCaffery J M, DeVries L, Farquhar M G. Proc Natl Acad Sci USA. 1999;96:6722–6727. doi: 10.1073/pnas.96.12.6722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.De Vries L, Mousli M, Wurmser A, Farquhar M G. Proc Natl Acad Sci USA. 1995;92:11916–11920. doi: 10.1073/pnas.92.25.11916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Balch W E, McCaffery J M, Plutner H, Farquhar M G. Cell. 1994;76:841–852. doi: 10.1016/0092-8674(94)90359-x. [DOI] [PubMed] [Google Scholar]
- 26.Hamm H E, Deretic D, Arendt A, Hargrave P A, Koenig B, Hofmann K P. Science. 1988;241:832–835. doi: 10.1126/science.3136547. [DOI] [PubMed] [Google Scholar]
- 27.Miura K, Hirai M, Kanai Y, Kurosawa Y. Genomics. 1996;34:181–186. doi: 10.1006/geno.1996.0263. [DOI] [PubMed] [Google Scholar]
- 28.Wall M A, Posner B A, Sprang S R. Structure (London) 1998;6:1169–1183. doi: 10.1016/s0969-2126(98)00117-8. [DOI] [PubMed] [Google Scholar]
- 29.Denker B M, Boutin P M, Neer E J. Biochemistry. 1995;34:5544–5553. doi: 10.1021/bi00016a028. [DOI] [PubMed] [Google Scholar]
- 30.Tanaka T, Kohno T, Kinoshita S, Mukai H, Itoh H, Ohya M, Miyazawa T, Higashijima T, Wakamatsu K. J Biol Chem. 1998;273:3247–3252. doi: 10.1074/jbc.273.6.3247. [DOI] [PubMed] [Google Scholar]
- 31.Sprang S R. Annu Rev Biochem. 1997;66:639–678. doi: 10.1146/annurev.biochem.66.1.639. [DOI] [PubMed] [Google Scholar]
- 32.Mixon M B, Lee E, Coleman D E, Berghuis A M, Gilman A G, Sprang S R. Science. 1995;270:954–960. doi: 10.1126/science.270.5238.954. [DOI] [PubMed] [Google Scholar]
- 33.Miura K, Kurosawa Y, Kanai Y. Biochem Biophys Res Commun. 1994;199:1388–1393. doi: 10.1006/bbrc.1994.1384. [DOI] [PubMed] [Google Scholar]
- 34.Coleman D E, Sprang S R. Biochemistry. 1998;37:14376–85. doi: 10.1021/bi9810306. [DOI] [PubMed] [Google Scholar]
- 35.Gilman A G. Annu Rev Biochem. 1987;56:615–649. doi: 10.1146/annurev.bi.56.070187.003151. [DOI] [PubMed] [Google Scholar]
- 36.Wilson B S, Komuro M, Farquhar M G. Endocrinology. 1994;134:233–244. doi: 10.1210/endo.134.1.8275939. [DOI] [PubMed] [Google Scholar]
- 37.Denker S P, McCaffery J M, Palade G E, Insel P A, Farquhar M G. J Cell Biol. 1996;133:1027–1040. doi: 10.1083/jcb.133.5.1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stow J L, Sabolic I, Brown D. Am J Physiol. 1991;261:F831–F840. doi: 10.1152/ajprenal.1991.261.5.F831. [DOI] [PubMed] [Google Scholar]
- 39.De Vries L, Farquhar M G. Trends Cell Biol. 1999;9:138–144. doi: 10.1016/s0962-8924(99)01515-9. [DOI] [PubMed] [Google Scholar]
- 40.Stow J L, de Almeida J B, Narula E J, Holtzman E J, Ercolani L, Ausiello D A. J Cell Biol. 1991;114:1113–1124. doi: 10.1083/jcb.114.6.1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tesmer J J, Berman D M, Gilman A G, Sprang S R. Cell. 1997;89:251–261. doi: 10.1016/s0092-8674(00)80204-4. [DOI] [PubMed] [Google Scholar]