Abstract
Gene regulatory circuits are often commonly shared between two closely related organisms. Our web tool iCR (identify Conserved target of a Regulon) makes use of this fact and identify conserved targets of a regulatory protein. iCR is a special refined extension of our previous tool PredictRegulon- that predicts genome wide, the potential binding sites and target operons of a regulatory protein in a single user selected genome. Like PredictRegulon, the iCR accepts known binding sites of a regulatory protein as ungapped multiple sequence alignment and provides the potential binding sites. However important differences are that the user can select more than one genome at a time and the output reports the genes that are common in two or more species. In order to achieve this, iCR makes use of Cluster of Orthologous Group (COG) indices for the genes. This tool analyses the upstream region of all user-selected prokaryote genome and gives the output based on conservation target orthologs. iCR also reports the Functional class codes based on COG classification for the encoded proteins of downstream genes which helps user understand the nature of the co-regulated genes at the result page itself. iCR is freely accessible at http://www.cdfd.org.in/icr/.
INTRODUCTION
Over last one and half decades, genomes of microorganisms have been sequenced at a highly accelerated pace. However, extracting useful information from such a large pool of genome data has become a major challenge of post genomics era. One approach to address this issue is to organize the large and complex genome into an ordered and manageable subsystem that can be tackled systematically. An important example of this approach is to study cellular processes and associated gene expression in terms of gene regulatory circuits. Each of these circuits contains a regulator and a list of its target sites (motifs) located upstream to a subset of genes that are being regulated (1–3). Such an approach will enable us to understand how the constituent genes of a genome come together to execute metabolic and physiological processes of a cell in response to a given regulator.
A large number of experimental and computational approaches are being attempted to understand how these genes come together to perform physiological function. The experimental approaches typically include microarray analysis of transcriptome (4,5). Subsequent to gathering the experimental data computational approaches are applied to search for common regulatory motifs and promoters present upstream to the up and down regulated genes and protein (6). Some of the computational tools like PHYLONET (7), BioProspector (8,9), Compare Prospector (9,10), MDscan (9,11), Motif Regressor (12), Bio Optimizer (13), PhyME (14) and so on are available for this purpose, but, most of these are either designed for eukaryotes or written to analyze the experimental data, such as micro array data, in terms of gene regulation.
An alternate approach could be to first select the regulator associated with a cellular process and then use computational approach to identify the potential target of regulatory protein which could then subsequently be followed up by experiments to validate the computationally identified targets. As a first step in this direction, we had previously proposed a tool called PredictRegulon, which finds targets of a regulatory protein in a genome based on limited set of known binding motif data (15). We have successfully used this tool to identify and validate the DtxR and IdeR targets in corynebacteria and mycobacteria, respectively (16,17). However an important limitation of Predictregulon was that it searches one genome at a time.
Carrying out simultaneous search in multiple genomes offers many advantages, most important among these are ability of such approach to reveal the conserved regulatory targets across the multiple related genomes. This would increase the confidence of experimental biologist in taking up experimental validation. Further it was also felt that if we could group the targets based on class of genes that is being regulated then we could provide the overall impact of the regulator on the physiology of the organism.
We describe here iCR (identify Conserved target of a Regulon), a web server tool, for identification of conserved high priority targets of a regulatory protein from heterologous sequence data of prokaryotes (which includes regulatory sequences of genes and their orthologs in other species) where the user can easily distinguish biologically important motifs from background noise based on their cross species conservation.
PROGRAM DESCRIPTION
iCR is a CGI based web application written in Perl and C language. It uses a Shannon relative entropy based profile search method, similar to what was used in PredictRegulon tool. This application can utilize the available experimental data on binding sites of a transcription regulatory protein (18–20) to identify the regulons of a given regulator in genomes of various phylogenetically related bacterial species.
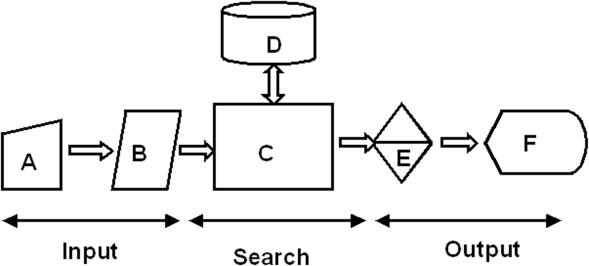
iCR is composed of three parts (Figure 1): (i) a front-end web interface for submitting the block aligned known binding motifs and for selection of species of choice; (ii) a search engine for scanning the upstream sequences; and (iii) a classification and reporting system for rendering the textual output produced by iCR into a meaningful grouping. Each of these components is discussed in detail in the help pages linked to the iCR home page. A brief description is being given here.
Figure 1.
Architecture of iCR. iCR is a CGI application which collects input from user using html forms (A). B represents a Perl script that gathers the input from A launches the Search Engine (C) which looks up genome sequences and their annotations (D), and returns the potential targets as an output which is further classified based on COG/Class or Genome. The classified output is returned as HTML output (F).
Input submission
iCR provides a web-based form for the input submission. The input form consists of two HTML pages. The first one accepts the sample motifs and the parameters defining the upstream region. On this page the known motifs can be copied either from sample input form or any authentic source and then be pasted in the web form in a block aligned fashion. The second page has a list of genomes organized in a taxonomically meaningful order for convenience in selection of multiple related species at a time and finally, the users need to specify the basis on which they want the predicted motifs to be grouped or classified on. The default or preferred option is Cluster of Orthologous Group (COG).
Search engine
Parameters accepted from the input forms are passed to a search engine which uses the Shannon relative entropy based profile scan method to scan the upstream sequences for regulatory motifs. This method is described in our previous paper PredictRegulon (15). However this analysis is carried out on multiple user selected genome and the results are compiled together. Since the complete COG data were not available for many of the genes of various genomes, we updated these data by running COGNITOR (21,22). Each COG selected represents the best hits to proteins from at least three lineages.
The output of the search result is classified and grouped based on one of the three options—orthology, function class code or genome. Classification based on orthology (default option) lists all the orthologous targets of a regulator together emphasizing the fact that these are conserved targets of a given regulon.
Output
All the predicted and classified target motifs are presented as HTML table. This table has following columns: COG name, Functional class code, Genome, motif score, motif, Gene id mentioned in NCBI's ptt table, ORF number and gene product. The program predicts a number of motifs, the blue background color shows the high scoring motifs above the cut-off value. The motifs with yellow background color depicts exact match to the known binding sites.
Example usage
To demonstrate the typical application of iCR's regulon assignments, we chose to use known LexA-binding sites from Bacillus subtilis as a query set. These sites were collected from PRODORIC (19). We then selected different species belonging to Fermicutes (Bacillales, Lactobacillales, Clostridia and Mollicutes) simultaneously for search. We obtained the result classified on COG in which DNA motifs upstream to lexA (COG1974), recA(COG0468), uvrB(COG0556), dinP(COG0389), rpsE(COG0098), rpsN(COG0098), rggD (COG0457) and so on were picked up in many species together and therefore they qualify for conserved targets of LexA regulon (Table 1). Lex A is known to autoregulates itself (23). recA gene has been experimentally shown to be part of LexA regulon in Escherichia coli as well as B.subtilis (23,24). Homologs of dinP have also been shown to be regulated by LexA protein in Bdellovibrio bacteriovorus (25). LexA protein has been reported to interact with the regulatory region of uvrB in B.subtilis (19). All these observations confirm that the program is capable of identifying significant and high priority targets of a given regulator successfully. Additionally the result also highlights many motifs upstream to hypothetical genes/ORFs. An experimental confirmation of interaction of these motifs to LexA, followed by a functional assay based on known processes involved with a given regulator, could shed more lights on function of these hypothetical genes.
Table 1.
Output of iCR showing the conserved targets of LexA regulon in Fermicutes
| COG | Class | Genome | Score | Position | Site | Gene | Synonym |
|---|---|---|---|---|---|---|---|
| COG1974 | K | NC_004193 | 4.6875 | −77 | AGAACGAGTGTTTG | lexA | OB1669 |
| COG1974 | K | NC_003030 | 4.77125 | −84 | AGAACATAAGTTTG | lexA | CAC1832 |
| COG1974 | K | NC_002745 | 4.88271 | −71 | CGAACAAATGTTTG | lexA | SA1174 |
| COG1974 | K | NC_004557 | 4.82946 | −80 | AGAACATAAGTTTG | lexA | CTC01298 |
| COG1974 | K | NC_003366 | 4.83493 | −70 | AGAACATAAGTTTG | lexA | CPE1161 |
| COG1974 | K | NC_002570 | 4.72756 | −77 | AGAACTTATGTTTG | lexA | BH2356 |
| COG1974 | K | NC_000964 | 4.81601 | −118 | CGAACCTATGTTTG | lexA | BSU17850 |
| COG1974 | K | NC_003923 | 4.88303 | −71 | CGAACAAATGTTTG | lexA | MW1226 |
| COG1974 | K | NC_003212 | 4.82162 | −79 | CGAACCTTTGTTTG | — | LIN1340 |
| COG1974 | K | NC_002758 | 4.88182 | −138 | CGAACAAATGTTTG | lexA | SAV1339 |
| COG1974 | K | NC_003210 | 4.81541 | −79 | CGAACCTTTGTTTG | — | LMO1302 |
| COG0468 | L | NC_002570 | 4.64423 | −121 | CGAATAAATGTTCG | recA | BH2383 |
| COG0468 | L | NC_003212 | 4.67474 | −138 | CGAATAAATGTTCG | recA | LIN1435 |
| COG0468 | L | NC_003210 | 4.66915 | −138 | CGAATAAATGTTCG | recA | LMO1398 |
| COG0468 | L | NC_003923 | 4.40442 | −143 | AGCACGTTTGTTCG | recA | MW1168 |
| COG0468 | L | NC_002758 | 4.40302 | −80 | AGCACGTTTGTTCG | recA | SAV1285 |
| COG0468 | L | NC_003030 | 4.90549 | −48 | AGAACAAATGTTCG | recA | CAC1815 |
| COG0468 | L | NC_003366 | 5.01207 | −34 | AGAACTTATGTTCG | recA | CPE1673 |
| COG0468 | L | NC_004461 | 4.42484 | −143 | AGTACGTTTGTTCG | — | SE0963 |
| COG0468 | L | NC_000908 | 4.18494 | −236 | TGAACTGTTGTATG | recA | MG339 |
| COG0468 | L | NC_002745 | 4.40405 | −143 | AGCACGTTTGTTCG | recA | SA1128 |
| COG0468 | L | NC_004557 | 4.9426 | −54 | AGAACAGATGTTCG | recA | CTC01289 |
| COG0556 | L | NC_000964 | 4.7767 | −122 | CGAACTTTAGTTCG | uvrB | BSU35170 |
| COG0556 | L | NC_003923 | 4.8228 | −105 | CGAACAAACGTTTG | uvrB | MW0720 |
| COG0556 | L | NC_002745 | 4.82248 | −105 | CGAACAAACGTTTG | uvrB | SA0713 |
| COG0556 | L | NC_003030 | 4.93204 | −29 | CGAACAAATGTTTG | uvrB | CAC0502 |
| COG0556 | L | NC_002758 | 4.82157 | −103 | CGAACAAACGTTTG | uvrB | SAV0758 |
| COG0556 | L | NC_004193 | 4.65391 | −69 | CGAATACTTGTTCG | — | OB2488 |
| COG0556 | L | NC_003212 | 4.62091 | −158 | CGAAAATATGTTCG | uvrB | LIN2632 |
| COG0556 | L | NC_003210 | 4.61721 | −160 | CGAAAATATGTTCG | uvrB | LMO2489 |
| COG0556 | L | NC_004461 | 4.90087 | −128 | CGAACAAATGTTTG | — | SE0541 |
| COG0389 | L | NC_003366 | 4.82409 | −26 | TGAACATATGTTTG | dinP | CPE1566 |
| COG0389 | L | NC_003923 | 4.77999 | −49 | GGAACACGTGTTCG | — | MW1251 |
| COG0389 | L | NC_002758 | 4.33641 | −6 | AGAACATTTGTTCT | — | SAV1364 |
| COG0389 | L | NC_002745 | 4.81919 | −49 | AGAACACGTGTTCG | — | SA1196 |
| COG0389 | L | NC_003210 | 4.72978 | −33 | AGAACGCTTGTTCG | — | LMO1975 |
| COG0389 | L | NC_004461 | 4.32424 | −75 | AGAACAAATGTTCT | — | SE1046 |
| COG0389 | L | NC_003212 | 4.73647 | −33 | AGAACGCTTGTTCG | — | LIN2082 |
| COG0389 | L | NC_004557 | 4.82946 | −40 | AGAACATAAGTTTG | — | CTC00437 |
| COG0389 | L | NC_000964 | 4.37402 | −68 | CGAACATAAGTTCT | yqjW | BSU23710 |
| COG0199 | J | NC_004368 | 4.29015 | −280 | TGAACGTATGTACG | — | GBS0071 |
| COG0199 | J | NC_002662 | 4.9713 | −280 | CGAACGTATGTTCG | rpsN | L0391 |
| COG0199 | J | NC_003028 | 4.22998 | −280 | TGAACGTATGTACG | — | SP0222 |
| COG0199 | J | NC_003098 | 4.22977 | −280 | TGAACGTATGTACG | rpsN | SPR0202 |
| COG0199 | J | NC_002737 | 4.41534 | −278 | CGAACGTATGTACG | rpsN | SPY0064 |
| COG0199 | J | NC_003485 | 4.41477 | −278 | CGAACGTATGTACG | rpsN | SPYM18_0065 |
| COG0199 | J | NC_004432 | 4.29794 | −140 | CGAAATTGTGTATG | — | MYPE10040 |
| COG0199 | J | NC_004070 | 4.41397 | −278 | CGAACGTATGTACG | rpsN.1 | SPYM3_0053 |
| COG0199 | J | NC_004116 | 4.2898 | −280 | TGAACGTATGTACG | — | SAG0071 |
| COG1396 | K | NC_003485 | 4.21446 | −8 | AGAAACCATGTTAG | — | SPYM18_0038 |
| COG1396 | K | NC_003923 | 4.32136 | −263 | GGAACAAGTGTACG | — | MW1228 |
| COG1396 | K | NC_004070 | 4.21434 | −8 | AGAAACCATGTTAG | — | SPYM3_0031 |
| COG1396 | K | NC_002570 | 4.4873 | −118 | GGAACGGGCGTTTG | — | BH0096 |
| COG1396 | K | NC_003028 | 4.47861 | −127 | TGAACAAATGTTGG | — | SP1115 |
| COG1396 | K | NC_002737 | 4.21453 | −8 | AGAAACCATGTTAG | — | SPY0037 |
| COG1396 | K | NC_004193 | 4.36271 | −253 | TGAACAGGAGTTAG | — | OB3501 |
| COG1396 | K | NC_003366 | 4.35319 | −58 | TGAACATTTGATTG | — | CPE2564 |
| COG0098 | J | NC_003028 | 4.38376 | −109 | AGAAGTGGTGTTCG | — | SP0227 |
| COG0098 | J | NC_004116 | 4.25066 | −110 | TGAAGTGGTGTTTG | rpsE | SAG0075 |
| COG0098 | J | NC_002737 | 4.23373 | −110 | TGAAGTGGTGTTTG | rpsE | SPY0069 |
| COG0098 | J | NC_004368 | 4.25082 | −110 | TGAAGTGGTGTTTG | rpsE | GBS0075 |
| COG0098 | J | NC_003098 | 4.38367 | −109 | AGAAGTGGTGTTCG | rpsE | SPR0206 |
| COG0098 | J | NC_004070 | 4.23345 | −110 | TGAAGTGGTGTTTG | rpsE | SPYM3_0057 |
| COG0098 | J | NC_004350 | 4.24208 | −109 | TGAAGTGGTGTTTG | rs5 | SMU.2009 |
| COG0098 | J | NC_003485 | 4.23361 | −110 | TGAAGTGGTGTTTG | rpsE | SPYM18_0069 |
| COG0457 | R | NC_004557 | 4.34686 | −240 | GGAAGAAGAGTTTG | — | CTC02554 |
| COG0457 | R | NC_002570 | 4.38934 | −268 | CGAAGCAACGTTTG | — | BH3054 |
| COG0457 | R | NC_004557 | 4.39536 | −233 | AGAACAATTGTATG | — | CTC01089 |
| COG0457 | R | NC_002745 | 4.37946 | −17 | AGAAATGAGGTTCG | — | SA1448 |
| COG0457 | R | NC_003923 | 4.3797 | −17 | AGAAATGAGGTTCG | — | MW1570 |
| COG0457 | R | NC_003098 | 4.47855 | −97 | TGAACAAATGTTGG | rggD | SPR1022 |
| COG0457 | R | NC_002758 | 4.3788 | −86 | AGAAATGAGGTTCG | — | SAV1620 |
Note: Gene, Synonym column is as per NCBI ptt table. Class codes—K involved in transcription, L in DNA replication, recombination and repair, J represents orthologs involved in translation, ribosomal structure and biogenesis and so on.
To test the sensitivity of the iCR predictions, we deleted two important and known binding motifs of LexA protein (present upstream to the dinB and uvrB in B.subtilis) from the input form and selected two species of Bacillales, B.subtilis and Bacillus holodurans. These two motifs were picked up on result page with blue background proving the reliability of predictions.
Certainly iCR results can serve as a useful starting point for molecular and cellular biologists for designing experiments to see the in vitro and in vivo effects of a regulatory protein in different systems.
CONCLUSION
To summarize, iCR is a web server that permits high throughput, detailed and fully automated prediction of potential binding targets of a regulatory protein in user selected prokaryotic species. iCR consists of 115 prokaryotic species arranged phylogenetically on the web interface. The first column on the result page, COG, is hyperlinked to NCBI and are fully navigable to allow users to have easy access to more related and descriptive information. The genome column shows the genome ID that is hyperlinked to a HTML page containing genome names corresponding to different IDs. For the user's convenience, functional class code column has also been linked to a page, which has a description of all the codes. iCR's strengths are in its free web accessibility, its comprehensiveness regarding choice of multiple species at a time, sorting of result based on COG and Class, and its interactive graphical interface.
Acknowledgments
Research in AR's laboratory is supported by grants from Council of Scientific and Industrial Research (CSIR) NMITLI, Department of Biotechnology, Department of Science and Technology, Govt. of India. S.R. and J.S. are supported by CSIR NMITLI Grant. V.V. is supported by UGC Research Fellowship and Y.S. is supported by CSIR Research Fellowship. Funding to pay the Open Access publication charges for this article was provided by Centre for DNA Fingerprinting and Diagnostics, Hyderabad.
Conflict of interest statement. None declared.
REFERENCES
- 1.Xing B., van der Laan M.J. A causal inference approach for constructing transcriptional regulatory networks. Bioinformatics. 2005;21:4007–4013. doi: 10.1093/bioinformatics/bti648. [DOI] [PubMed] [Google Scholar]
- 2.Hershberg R., Yeger-Lotem E., Margalit H. Chromosomal organization is shaped by the transcription regulatory network. Trends Genet. 2005;21:138–142. doi: 10.1016/j.tig.2005.01.003. [DOI] [PubMed] [Google Scholar]
- 3.Balazsi G., Barabasi A.L., Oltvai Z.N. Topological units of environmental signal processing in the transcriptional regulatory network of Escherichia coli. Proc. Natl Acad. Sci. USA. 2005;102:7841–7846. doi: 10.1073/pnas.0500365102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ren J., Prescott J.F. Analysis of virulence plasmid gene expression of intra-macrophage and in vitro grown Rhodococcus equi ATCC 33701. Vet. Microbiol. 2003;94:167–182. doi: 10.1016/s0378-1135(03)00099-3. [DOI] [PubMed] [Google Scholar]
- 5.Rodriguez G.M., Voskuil M.I., Gold B., Schoolnik G.K., Smith I. ideR, an essential gene in Mycobacterium tuberculosis: role of IdeR in iron-dependent gene expression, iron metabolism, and oxidative stress response. Infect. Immun. 2002;70:3371–3381. doi: 10.1128/IAI.70.7.3371-3381.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lin L.H., Lee H.C., Li W.H., Chen B.S. Dynamic modeling of cis-regulatory circuits and gene expression prediction via cross-gene identification. BMC Bioinformatics. 2005;6:258. doi: 10.1186/1471-2105-6-258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang T., Stormo G.D. Identifying the conserved network of cis-regulatory sites of a eukaryotic genome. Proc. Natl Acad. Sci. USA. 2005;102:17400–17405. doi: 10.1073/pnas.0505147102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu X., Brutlag D.L., Liu J.S. BioProspector: discovering conserved DNA motifs in upstream regulatory regions of co-expressed genes. Pac. Symp. Biocomput. 2001:127–138. [PubMed] [Google Scholar]
- 9.Liu Y., Wei L., Batzoglou S., Brutlag D.L., Liu J.S., Liu X.S. A suite of web-based programs to search for transcriptional regulatory motifs. Nucleic Acids Res. 2004;32:W204–W207. doi: 10.1093/nar/gkh461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu Y., Liu X.S., Wei L., Altman R.B., Batzoglou S. Eukaryotic regulatory element conservation analysis and identification using comparative genomics. Genome Res. 2004;14:451–458. doi: 10.1101/gr.1327604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu X.S., Brutlag D.L., Liu J.S. An algorithm for finding protein–DNA binding sites with applications to chromatin-immunoprecipitation microarray experiments. Nat. Biotechnol. 2002;20:835–839. doi: 10.1038/nbt717. [DOI] [PubMed] [Google Scholar]
- 12.Conlon E.M., Liu X.S., Lieb J.D., Liu J.S. Integrating regulatory motif discovery and genome-wide expression analysis. Proc. Natl Acad. Sci. USA. 2003;100:3339–3344. doi: 10.1073/pnas.0630591100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jensen S.T., Liu J.S. BioOptimizer: a Bayesian scoring function approach to motif discovery. Bioinformatics. 2004;20:1557–1564. doi: 10.1093/bioinformatics/bth127. [DOI] [PubMed] [Google Scholar]
- 14.Sinha S., Blanchette M., Tompa M. PhyME: a probabilistic algorithm for finding motifs in sets of orthologous sequences. BMC Bioinformatics. 2004;5:170. doi: 10.1186/1471-2105-5-170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yellaboina S., Seshadri J., Kumar M.S., Ranjan A. PredictRegulon: a web server for the prediction of the regulatory protein binding sites and operons in prokaryote genomes. Nucleic Acids Res. 2004;32:W318–W320. doi: 10.1093/nar/gkh364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yellaboina S., Ranjan S., Chakhaiyar P., Hasnain S.E., Ranjan A. Prediction of DtxR regulon: identification of binding sites and operons controlled by Diphtheria toxin repressor in Corynebacterium diphtheriae. BMC Microbiol. 2004;4:38. doi: 10.1186/1471-2180-4-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Prakash P., Yellaboina S., Ranjan A., Hasnain S.E. Computational prediction and experimental verification of novel IdeR binding sites in the upstream sequences of Mycobacterium tuberculosis open reading frames. Bioinformatics. 2005;21:2161–2166. doi: 10.1093/bioinformatics/bti375. [DOI] [PubMed] [Google Scholar]
- 18.Salgado H., Santos-Zavaleta A., Gama-Castro S., Millan-Zarate D., Diaz-Peredo E., Sanchez-Solano F., Perez-Rueda E., Bonavides-Martinez C., Collado-Vides J. RegulonDB (Version 3.2): transcriptional regulation and operon organization in Escherichia coli K-12. Nucleic Acids Res. 2001;29:72–74. doi: 10.1093/nar/29.1.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Munch R., Hiller K., Barg H., Heldt D., Linz S., Wingender E., Jahn D. PRODORIC: prokaryotic database of gene regulation. Nucleic Acids Res. 2003;31:266–269. doi: 10.1093/nar/gkg037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ishii T., Yoshida K., Terai G., Fujita Y., Nakai K. DBTBS: a database of Bacillus subtilis promoters and transcription factors. Nucleic Acids Res. 2001;29:278–280. doi: 10.1093/nar/29.1.278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tatusov R.L., Galperin M.Y., Natale D.A., Koonin E.V. The COG database: a tool for genome-scale analysis of protein functions and evolution. Nucleic Acids Res. 2000;28:33–36. doi: 10.1093/nar/28.1.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tatusov R.L., Natale D.A., Garkavtsev, IV, Tatusova T.A., Shankavaram U.T., Rao B.S., Kiryutin B., Galperin M.Y., Fedorova N.D., Koonin E.V. The COG database: new developments in phylogenetic classification of proteins from complete genomes. Nucleic Acids Res. 2001;29:22–28. doi: 10.1093/nar/29.1.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Little J.W., Mount D.W., Yanisch-Perron C.R. Purified lexA protein is a repressor of the recA and lexA genes. Proc. Natl Acad. Sci. USA. 1981;78:4199–4203. doi: 10.1073/pnas.78.7.4199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Groban E.S., Johnson M.B., Banky P., Burnett P.G., Calderon G.L., Dwyer E.C., Fuller S.N., Gebre B., King L.M., Sheren I.N., et al. Binding of the Bacillus subtilis LexA protein to the SOS operator. Nucleic Acids Res. 2005;33:6287–6295. doi: 10.1093/nar/gki939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Campoy S., Salvador N., Cortes P., Erill I., Barbe J. Expression of canonical SOS genes is not under LexA repression in Bdellovibrio bacteriovorus. J. Bacteriol. 2005;187:5367–5375. doi: 10.1128/JB.187.15.5367-5375.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]