Abstract
GeneAlign is a coding exon prediction tool for predicting protein coding genes by measuring the homologies between a sequence of a genome and related sequences, which have been annotated, of other genomes. Identifying protein coding genes is one of most important tasks in newly sequenced genomes. With increasing numbers of gene annotations verified by experiments, it is feasible to identify genes in the newly sequenced genomes by comparing to annotated genes of phylogenetically close organisms. GeneAlign applies CORAL, a heuristic linear time alignment tool, to determine if regions flanked by the candidate signals (initiation codon-GT, AG-GT and AG-STOP codon) are similar to annotated coding exons. Employing the conservation of gene structures and sequence homologies between protein coding regions increases the prediction accuracy. GeneAlign was tested on Projector dataset of 491 human–mouse homologous sequence pairs. At the gene level, both the average sensitivity and the average specificity of GeneAlign are 81%, and they are larger than 96% at the exon level. The rates of missing exons and wrong exons are smaller than 1%. GeneAlign is a free tool available at http://genealign.hccvs.hc.edu.tw.
INTRODUCTION
Accurate prediction of gene structures, precise exon–intron boundaries, is an essential step in analysis of genomic sequences. Despite numerous developments of useful tools, no programs can predict all the protein coding genes perfectly (1). Single-genome predictors which predict gene structures by using one genomic sequence, e.g. GENSCAN (2), have been successfully used at the prediction of newly sequenced genomes. However, the best accuracy is achieved by the spliced alignment of full-length cDNAs or comprehensive expressed sequences tags (ESTs) (3). Sim4, Spidey and GMAP (4–6) belong to the latter class. Due to incomplete sequence information of a transcriptome, a completely accurate prediction of the corresponding genome is still an existing challenge. With more and more genomes being sequenced, the comparative approaches become more feasible. Several programs, e.g. TWINSCAN (7), SGP2 (8), SLAM (9) and EXONALIGN (10), have been developed to compare genomes of related organisms. In addition to the comparative analysis between genomes, evidences from related organisms have been employed in the comparative approaches. The programs, GeneSeqer (3), GeneWise (11) and Projector (12), have been developed to utilize evidences of cDNAs/ESTs, known proteins and known annotations of related organisms, respectively, to help gene prediction. Recently, ExonHunter (13) and JIGSAW (14) have been developed to further increase the accuracy for gene prediction by integrating multiple sources of information including multiple genomic sequences, protein databases, cDNAs/ESTs of related organisms and the output of various gene predictors.
This paper presents a web tool, GeneAlign, for protein coding gene prediction. Same as Projector, GeneAlign employs annotated genes of one organism to predict the homologous genes of another organism. GeneAlign integrates signal detectors with CORAL (10) to efficiently align annotated coding exons with queried sequences. CORAL, a heuristic alignment program, aligns coding regions between two phylogenetically close organisms in linear time. The approach applied by GeneAlign can identify distinctive features of well conserved gene structures and protein coding sequences between phylogenetically close organisms. GeneAlign assumes the conservation of the exon–intron structures, but it can also align some exons which differ by events of exon-splitting and exon-fusion. In addition, GeneAlign has an explicit procedure for detecting micro-exons, which is usually a difficult task for eukaryotic gene prediction (15). Despite their small sizes, experimental studies support that small exons are usually conserved between organisms (16). A procedure for identifying micro-exons has been developed by Volfovsky et al. (17), and has been applied in a large scale study. GMAP (6) furthers this work by integrating the detection procedure into the framework of a cDNA-genomic alignment program. GeneAlign looks for potential micro-exons with the appropriate boundaries and computes the optimal alignments for these potential micro-exons and corresponding annotated exons. GeneAlign can predict gene structure by employing a fairly diverged annotated genome with conserved gene structure. Here, we show that GeneAlign performs well in identifying coding exons; specifically the rates of missing exons and wrong exons are both low.
MATERIALS AND METHODS
GeneAlign accepts 2 nt sequences of homologous genes and the known gene annotation of one of these two genes as inputs and predicts the coding exon positions in another sequence according to the known gene annotation. The major components of GeneAlign for annotation-genome mapping and alignment include: (i) signal filtrations, (ii) applying CORAL to measure the sequence homologies following candidate signals for generating approximate gene structures and (iii) recognition of micro-exons.
Signal filtrations
Splice sites are the most powerful signals for gene prediction, accurate modeling splice sites can improve the accuracy of gene prediction (1). To model the conserved gene structures of homologous genes, GeneAlign measures sequence homologies between annotated exons of one sequence and downstream/upstream to the potential splice acceptors/donors of another sequence. For the queried sequence, GeneAlign firstly obtains a set of candidate signals, splice acceptors/donors, according to signal scores calculated by GeneSplicer (18), the signal prediction program. The GeneSplicer, combined the Markov modeling techniques with a decision tree method (maximal dependence decomposition), detects splice sites in various eukaryotic genomes. The cutoff scores of candidate signals were set at −5 (default values) for splice acceptors and donors. The false negative (FN) and the false positive (FP) rates are respectively less than 2 and 10% for both acceptors and donors, showing that only 2% of true signals are missed and nearly 90% of wrong signals are filtered out. The GeneSplicer can efficiently filter out many false splice signals but failed to remove false signals resulting from highly degenerate and unspecific nature. CORAL (10) is integrated to measure sequence homologies between potential regions marked by splice signals and annotated exons.
COding Region ALignment—CORAL
CORAL is developed on the basis of the conservation of coding regions. Most of coding regions among organisms are conserved at the amino acid level, suggesting that the hamming distance of two segments with an optimal alignment is low. Relative to SPA (19), a probabilistic filtration method is built to efficiently find an ill-positioned pair. The ill-positioned pair is a less than optimal alignment, which is supposed to result from a shifting mutation and can be solved by inserting a gap with a length of a multiple of three. A local optimal solution is used to obtain a significant alignment when an ill-positioned pair is detected and to determine the possible position and length for the inserted gap. Considering that the nucleotide sequences of the translated regions are well conserved in the first and second positions of a codon and maybe less conserved in the third nucleotide of a codon, we utilized 3 nt spread out in the pattern XXO (where the X indicated ‘absolute matching’ and the O meant ‘don't care’) to serve as the basis of alignment. CORAL employs the probabilistic analysis and the local optimal solution to efficiently align sequences by sliding windows and, thus, obtains a near optimal alignment in linear time. The detail for the concept of CORAL can be referred to Hsieh et al. (10).
Gene Structure Alignment—GeneAlign
After signal filtrations by GeneSplicer, the queried sequences and annotated exons are aligned from 5′ to 3′. GeneAlign is designed for detecting multi-exons genes. The coding exons are divided into three categories according to their location in the coding region, initial exon (initiation codon-GT, first coding exon of a gene), internal exon (AG-GT) and terminal exon (AG-stop codon, last coding exon of a gene). The alignments by CORAL are processed from the splice acceptors by aligning the first annotated internal exons with regions following the candidate splice acceptors. CORAL stops aligning when the alignment score drops significantly. The aligned subsequence is predicted as a candidate exon when the alignment score (≥50%) and aligned sequence length (≥30 bp) are greater than the thresholds, which have been determined empirically. Candidate splice acceptors and the next annotated exons are examined subsequently to search for meaningful alignments. For each aligned segment, the downstream boundary is delimited by an admissible candidate splice donor. A series of aligned segments is ended at the annotated terminal exon and delimited by a stop codon, e.g. TAG, TGA and TAA. The aforementioned process is repeated from 3′ to 5′, from the last internal exons aligning with the regions following the candidate splice donors, and is ended at the annotated initial exon with an initiation codon (ATG). This procedure retrieves possible missing exons resulted from underestimation of splice acceptors by GeneSplicer, a single intron insertion/deletion to one of the exon pair, and frameshifts at the 5′ end of exon pairs. If the annotated exons cannot be mapped to the queried sequence, a lower threshold of the alignment score, e.g. 35%, will be reset, and the corresponding region is searched again. Although GeneAlign is designed to predict multi-exons genes, it can also predict single-exon genes with same structures by aligning the annotated exons with regions following the candidate translation initiation sites, which are predicted using a weight matrix model (WMM) (20).
Recognition of micro-exons
The micro-exons, smaller than 30 bp in length, are frequently encountered in the eukaryotic genomes (6,17); however, they cannot be detected by applying CORAL. Micro-exons in the annotated genes are processed by an additional procedure. Our method assumes that micro-exons are flanked by canonical boundaries. The sequence alignment is processed by a standard dynamic programming algorithm in order to compute the optimal alignment. The sequence homologies are assessed at the amino acid level by translating corresponding segments according to annotated translational reading frame and the genetic code. The resulting peptide segments are then aligned by the BLOSUM 62 substitution matrix (21). An amino acid match is defined as BLOSUM score larger than zero. A micro-exon is predicted only if its sequence identity larger than 50% and is flanked by canonical boundaries.
The alignment only applied in a specific region of nucleotide sequence corresponding to the position of micro-exon in the annotated gene. In addition, a large splice site score (e.g. score larger than zero) and an appropriate potential micro-exon length are required to offset the high probability of an exact match by chance. The length of an appropriate potential micro-exon differs with that of the corresponding annotated exon by a multiple of three and smaller than three codons insertion/deletion. When the aforementioned criteria are met, the program tests potential micro-exons for the alignment until an alignment with sequence identity larger than 50%.
RESULTS
GeneAlign applies CORAL based on the codon identity to efficiently find the partner exons to those of related known genes. The parameters are optimized by the IMOG dataset (8) of 15 homologous human–mouse gene pairs (10). The testing dataset is the Projector dataset (12) which collects 491 homologous human–mouse gene pairs not overlapping with the training set. The average number of exons per gene in the test set is 8.8 exons. Forty four percent of these gene pairs (216 out of 491) have the identical number of coding exons and the identical coding sequence length. Fifty one percent (249 out of 491) have identical exons number but differ in coding sequence length. Five percent (26 out of 491) have different number of exons. The human–mouse gene pairs share 14 initial micro-exons and 15 terminal micro-exons. They differ in the numbers of internal micro-exons that mouse has 18 and human has 19 internal micro-exons.
The performance of GeneAlign was evaluated separately by the accuracy of predictions for human and mouse genes and was compared with the outputs from Projector and GeneWise (11). The Projector program predicts gene structures by using the annotated genes of a related organism, which is the same with GeneAlign. The GeneWise program, predicting gene structures by using the known proteins of a related organism, serves as a benchmark (12). The sets of genes predicted by Projector and GeneWise were retrieved from the Projector web sever (http://www.sanger.ac.uk/Software/analysis/projector). We measured the performance in terms of sensitivity and specificity at both the exon and the gene levels. The results are summarized in Table 1. These results show that the predictions obtained by GeneAlign are accurate at both levels. The rates of missing exons and wrong exons are smaller than 1%. The prediction statistics of micro-exons are summarized in Table 2. The prediction accuracies of initial, internal and terminal micro-exons are respectively 96, 92 and 93%. Although GeneAlign misses more micro-exons than Projector, it predicts much less wrong micro-exons. The wrongly predicted micro-exons affect the performance of Projector at the gene level.
Table 1.
Prediction accuracy on the Projector dataset
| Program | Gene level* (%) | Exon level* (%) | ||||
|---|---|---|---|---|---|---|
| Sn | Sp | Sn | Sp | ME | WE | |
| Human gene prediction | ||||||
| GeneWise | 61.91 | 61.91 | 92.56 | 93.60 | 1.50 | 0.32 |
| Projector | 51.32 | 51.32 | 93.78 | 86.99 | 0.88 | 8.59 |
| GeneAlign | 82.28 | 82.28 | 96.65 | 97.12 | 0.74 | 0.32 |
| Mouse gene prediction | ||||||
| GeneWise | 60.49 | 60.49 | 93.13 | 93.39 | 1.18 | 0.28 |
| Projector | 58.45 | 58.45 | 94.55 | 90.35 | 0.47 | 4.55 |
| GeneAlign | 79.23 | 79.23 | 96.63 | 96.39 | 0.49 | 0.58 |
*The measures of sensitivity (Sn) and specificity (Sp) are respectively Sn= TP/(TP+ FN) and Sp= TP/(TP + FP). ME (missing exons) is the proportion of annotated exons not overlapped by any predicted exons, whereas WE (wrong exons) is the proportion of predicted exons not overlapped by any annotated exons.
Table 2.
Prediction accuracy on micro-exons of the Projector dataset
| Program | No. of micro-exons* | ||
|---|---|---|---|
| Accurate exons | Missing exons | Wrong exons | |
| Human micro-exon prediction | |||
| GeneWise | 22 | 25 | 2 |
| Projector | 45 | 1 | 339 |
| GeneAlign | 45 | 2 | 5 |
| Mouse micro-exon prediction | |||
| GeneWise | 23 | 22 | 3 |
| Projector | 47 | 0 | 170 |
| GeneAlign | 44 | 3 | 9 |
*The accuracy of identifying micro-exons was evaluated by the number of accurately predicted exons, missing exons and wrong exons. An exon is accurately predicted only when both boundaries are correct. Missing exons are annotated exons not overlapped with predicted exons. Wrong exons are predicted exons not overlapped by any annotated exons. In the Projector dataset, there are 48 and 47 micro-exons in human and mouse genes, respectively.
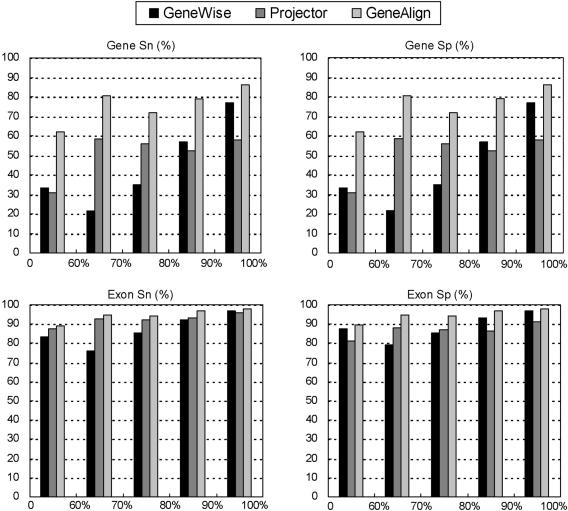
In order to study the effects of sequence homology on the performance of prediction accuracy, 491 homologous pairs were stratified into five classes with amino acid identities between two encoded proteins ranging from <60, 60–70, 70–80, 80–90 and 90–100% (Figure 1). The overall identities (amino acid identities) between two protein sequences encoded by the homologous gene pair were calculated by a standard dynamic programming algorithm. There are respectively 21, 23, 59, 154, 234 pairs in each class. Figure 1 shows that the performance of the three programs exhibits a strong dependence on the amino acid identities. GeneAlign, integrating a good splice signal detector and CORAL, can model the conservation of exon boundaries and the encoded amino acid sequences, and thus performs well in all classes of sequence homologies. Nevertheless, GeneAlign misses some exons with widely different gene structures for structure conservation is a pre-requested assumption. The missing and wrong exons predicted by GeneAlign were analyzed more in detail. Some of the wrongly predicted exons display high degree sequence conservation with annotated exons and the lengths are multiple of three. It is possible that some of these wrongly predicted exons may be expressed. In addition, some of the missing exons result from lack of partner exon annotations. The possibility of missing exons present in rare alternative splice forms in one of the human and mouse gene pair cannot be excluded. The set of genes predicted by GeneAlign can be obtained at http://genealign.hccvs.hc.edu.tw/about_genealign.htm.
Figure 1.
Comparisons of the correlation between sequence homology and the prediction performance of the GeneWise, Projector and GeneAlign. The gene pairs of Projector dataset were sorted into five classes by their amino acid identities (<60, 60–70, 70–80, 80–90 and 90–100%), and the performance was calculated for each class. The amino acid identities were obtained by using a standard dynamic programming algorithm to calculate the identities between two protein sequences encoded in each homologous gene pair. The measures of sensitivity (Sn) and specificity (Sp) are respectively Sn = TP/(TP + FN) and Sp = TP/(TP + FP).
WEB SERVER DESCRIPTION
Input
The input consists of 2 nt sequences in FASTA format and one known gene annotation in ‘General Feature Format’ (GFF) or ‘Gene Transfer Format’ (GTF). Examples and a detail description are available at http://genealign.hccvs.hc.edu.tw/genealign_help.htm. The maximal length of the sequences submitted to the web server is 200 kb. In the current version, known genes annotated on the mouse/human genome are applied to predict human/mouse genes. The nucleotide sequences for the prediction can be obtained by mapping the known genes of one organism to their corresponding locations within the genome of another organism using the BLAST programs. The pair of corresponding genome sequences and the known gene annotation are uploaded to the web server as inputs to GeneAlign, which would predict genes in the queried sequence according to the known genes of the corresponding genome sequence.
Output
The output of GeneAlign contains a prediction result in GFF and the alignments of predicted exons. In GFF format, each predicted exon is presented on one line with eight fields. These fields include a sequence name for prediction, the gene prediction program name, the feature type (CDS), the start and end positions of the predicted exon, the identities generated by CORAL, the forward or reverse strand and the reading frame. Additionally, if the input queried sequence contains genome position, the results can be explored further on the UCSC genome browser (22). The UCSC genome browser provides an excellent environment for comparing various information sources.
Acknowledgments
Funding to pay the Open Access publication charges for this article was provided by the National Science Council of the Republic of China (NSC 93-3112-B-007-008 and NSC 94-3112-B-007-002).
Conflict of interest statement. None declared.
REFERENCES
- 1.Brent M.R., Buigo R. Recent advances in gene structure prediction. Curr. Opin. Struct. Biol. 2004;14:264–272. doi: 10.1016/j.sbi.2004.05.007. [DOI] [PubMed] [Google Scholar]
- 2.Burge C., Karlin S. Prediction of complete gene structures in human genomic DNA. J. Mol. Biol. 1997;268:78–94. doi: 10.1006/jmbi.1997.0951. [DOI] [PubMed] [Google Scholar]
- 3.Brendel V., Xing L., Zhu W. Gene structure prediction from consensus spliced alignment of multiple ESTs matching the same genomic locus. Bioinformatics. 2004;20:1157–1169. doi: 10.1093/bioinformatics/bth058. [DOI] [PubMed] [Google Scholar]
- 4.Florea L., Hartzell G., Zhang Z., Rubin G.M., Miller W. A computer program for aligning a cDNA sequence with a genomic DNA sequence. Genome Res. 1998;8:967–974. doi: 10.1101/gr.8.9.967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wheelan S.J., Church D.M., Ostell J.M. Spidey: a tool for mRNA-to-genomic alignments. Genome Res. 2001;11:1952–1957. doi: 10.1101/gr.195301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wu T.D., Watanabe C.K. GMAP: a genomic mapping and alignment program for mRNA and EST sequences. Bioinformatics. 2005;21:1859–1875. doi: 10.1093/bioinformatics/bti310. [DOI] [PubMed] [Google Scholar]
- 7.Korf I., Flicek P., Duan D., Brent M.R. Integrating genomic homology into gene structure prediction. Bioinformatics. 2001;17:140–148. doi: 10.1093/bioinformatics/17.suppl_1.s140. [DOI] [PubMed] [Google Scholar]
- 8.Parra G., Agarwal P., Abril J.F., Wiehe T., Fickett J.W., Guigó R. Comparative gene prediction in human and mouse. Genome Res. 2003;13:108–117. doi: 10.1101/gr.871403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Alexandersson M., Cawley S., Pachter L. SLAM: cross-species gene finding and alignment with a generalized pair hidden Markov model. Genome Res. 2003;13:496–502. doi: 10.1101/gr.424203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hsieh S.J., Lin C.Y., Chung Y.S., Tang C.Y. Comparative exon prediction based on heuristic coding region alignment. Proceedings of 8th International Symposium on Parallel Architectures, Algorithms and Networks (ISAPN 2005); Las Vegas, Nevada, USA: IEEE Computer Society Press; 2005. pp. 14–19. [Google Scholar]
- 11.Birney E., Clamp M., Durbin R. GeneWise and Genomewise. Genome Res. 2004;14:988–995. doi: 10.1101/gr.1865504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Meyer I.M., Durbin R. Gene structure conservation aids similarity based gene prediction. Nucleic Acids Res. 2004;32:776–783. doi: 10.1093/nar/gkh211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brejova B., Brown D.G., Li M., Vinar T. ExonHunter: a comprehensive approach to gene finding. Bioinformatics. 2005;21:57–65. doi: 10.1093/bioinformatics/bti1040. [DOI] [PubMed] [Google Scholar]
- 14.Allen J.E., Salzberg S.L. JIGSAW: integration of multiple sources of evidence for gene prediction. Bioinformatics. 2005;21:3596–3603. doi: 10.1093/bioinformatics/bti609. [DOI] [PubMed] [Google Scholar]
- 15.Mathe C., Sagot M.F., Schiex T., Rouze P. Current methods of gene prediction, their strengths and weaknesses. Nucleic Acids Res. 2002;30:4103–4117. doi: 10.1093/nar/gkf543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Black D.L. Protein diversity from alternative splicing: a challenge for bioinformatics and post-genome biology. Cell. 2000;103:367–370. doi: 10.1016/s0092-8674(00)00128-8. [DOI] [PubMed] [Google Scholar]
- 17.Volfovsky N., Haas B.J., Salzberg S.L. Computational discovery of internal micro-exons. Genome Res. 2003;13:1214–1221. doi: 10.1101/gr.677503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pertea M., Lin X., Salzberg S.L. GeneSplicer: a new computational method for splicer site prediction. Nucleic Acids Res. 2001;29:1185–1190. doi: 10.1093/nar/29.5.1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shen S.Y., Yang J., Yao A., Hwang P. Super pairwise alignment (SPA): an efficient approach to global alignment for homologous sequences. J. Comp. Biol. 2002;9:477–486. doi: 10.1089/106652702760138574. [DOI] [PubMed] [Google Scholar]
- 20.Staden R. Computer methods to locate signals in nucleic acid sequences. Nucleic Acids Res. 1984;12:505–519. doi: 10.1093/nar/12.1part2.505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Henikoff S., Henikoff J.G. Amino acid substitution matrices from protein blocks. Proc. Natl Acad. Sci. USA. 1992;89:10915–10919. doi: 10.1073/pnas.89.22.10915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Karolchik D., Baertsch R., Diekhans M., Furey T.S., Hinrichs A., Lu Y.T., Roskin K.M., Schwartz M., Sugnet C.W., Thomas D.J., et al. The UCSC Genome Browser Database'. Nucleic Acids Res. 2003;31:51–54. doi: 10.1093/nar/gkg129. [DOI] [PMC free article] [PubMed] [Google Scholar]