Abstract
Alternative transcript diversity manifests itself a prime cause of complexity in higher eukaryotes. The Alternative Splicing Graph Server (ASGS) is a web service facilitating the systematic study of alternatively spliced genes of higher eukaryotes by generating splicing graphs for the compact visual representation of transcript diversity from a single gene. Taking a set of transcripts in General Feature Format as input, ASGS identifies distinct reference and variable exons, generates a transcript splicing graph, an exon summary, splicing events classification and a single line graph to facilitate experimental analysis. This freely available web service can be accessed at http://asgs.biolinfo.org.
INTRODUCTION
The discovery of the intron simultaneously in 1977 (1) by the groups of Sharp and Roberts, has been summarized by Gilbert (2) in 1978 as ‘genes in pieces’ for eukaryotes. The combinatorial possibilities of transcript generation from a limited number of genes is now attributed to alternative splicing (AS) of exons as a means to explore the protein landscape. Until recently, AS was considered a sporadic occurrence in an otherwise orderly world of one gene leading to one protein product. However, it is now established that AS is ubiquitous and a major contributor to the complexity of higher organisms, with almost 80% of human genes (20 000–25 000) estimated to be alternatively spliced, with each gene generating multiple mRNA products (3). Stetefeld and Ruegg (4) estimate that almost 50% of eukaryotic genes are alternatively spliced. AS changes the structure of transcripts, which has the potential to dramatically increase the functional diversity of encoded gene products and allow mRNA isoforms to be differentially regulated in disparate biological processes (5). Furthermore, its disruption is associated with many diseases, such as cardiovascular, cancer and neurodegenerative disorders. Given the widespread appearance and functional diversity, understanding the mechanism and regulation of AS is a major goal of modern biological and biomedical research.
As more eukaryotic genomes are sequenced and annotated, several databases dedicated to AS are now available (7–11), leading to genome-wide computational analysis, reviewed by Lee and Wang (12). Although AS databases give an insight into the amount of AS, they do not provide any visual representation and classification of the types of AS events occurring (12). As the number of transcripts per gene increases, it has become increasingly difficult to identify branch points and systematically analyse and classify AS events. Directed acyclic graphs were used by Modrek and Lee (13) for EST analysis, with the genomic DNA sequence as reference. Pevzner and co-workers (14) first used de Bruijn graphs to depict the transcripts alone, without referring to the genomic DNA sequence, where the maximum common sub-sequences between transcripts were condensed into nodes and the variable regions connected by edges. Such an approach has been used to generate the Alternative Splicing Gallery (ASG) resource (7).
Our approach has been to use directed acyclic splicing graphs, without a genomic DNA sequence as reference and defining exons as nodes, interconnected by introns as edges, with paths through the splicing graph representing the transcripts; such a schema was applied to the Drosophila melanogaster genome (8), to the DEDB data resource. Here, the first transcript served as a reference sequence to generate splicing graphs, with automatic rule-based classification of splicing events. The use of exons and introns as nodes and edges, respectively, has the intuitive advantage of biological interpretation.
The ease of representing a set of transcripts as a compact graphic is provided by the Alternative Splicing Graph server (ASGS), a web service for generating the splicing graph, with automatic ruled-based classification of AS events, to facilitate transcriptome analysis. We have also methodologically enhanced our earlier approach (8), to identify the most conserved exons as distinct, with the rest classified as variable exons, independent of the first transcript provided. Splicing junctions are often validated by RT–PCR, to verify gene models predicted by gene finding programs, exemplified by Eyras and co-workers (15) for the chicken genome. For this purpose, the collapsed splicing graph form, the ‘single line graph,’ is also provided by our server, which gives a comprehensive view of all splicing junctions in a single gene.
OVERVIEW OF WEB SERVICES
The ASGS web service provides dynamic image generation of splicing graphs by parsing and identifying distinct reference exons and their variants. The user can submit the transcript information for a particular gene in the General Feature Format (GFF) (16). Alternatively, a GFF file can be uploaded to the server. Links to utilities that convert other file formats to GFF are provided from the ‘create’ web page. The software is designed based on the MVC (Model-View-Controller) architecture, described elsewhere in detail (17) with design patterns, completely written in java, and deployed using the Java system application server, to harness the computing power on the client side.
ASGS aims to provide the global AS research community with free web services to visualize alternative transcript diversity. ASGS fulfils these collective aims using a three-step process showing individual transcript representation, splicing graph and single line splice pattern to facilitate RT–PCR experimental analysis. This visual representation is followed by a detailed listing of distinct and variable exons and the transcripts they occur in as well as an AS event classification. The splicing graphs are created and displayed as high-resolution downloadable images, for local analysis, publication and presentation. A detailed description of transcript input required for visualization and the output has been provided on the online help page. Useful links to AS resources and a list of references is also available at the ASGS website.
MATERIALS AND METHODS
Data input
As primary data source, ASGS uses input lines based on the GFF standard file format. Each GFF input line has nine required fields that must be tab-separated. To create splicing graphs, the user needs to first create a GFF file containing sequence information organized as a series of exon positional information (13). Different biological sequence tools are freely available to convert other formats to GFF. Data from the user in GFF is converted into the transcript input model and passes to the server side program for dynamic image generation. Our web application extracts gene structure information such as the transcript location and the start and end positions of each exon that make up the transcript, which are then parsed out and checked for consistency.
In order that transcript information is useful to the gene prediction and genome annotation community, we have designed a single line graph, of the type that is seen in molecular biology text books, by merging all the variant exons of distinct reference exons leading to a one-line summary transcript diversity diagram. This one-line graph is what an experimentalist will be interested in, for the AS events of a specific gene, as it suggests sequences for primer design across splice junctions. Further, AS events such as intron-retention are clearly seen in the single line graph.
ASGS also transforms any anti-sense transcripts to the sense direction before calling the transcript model and provides a detailed list of classification events to help experimental verification of AS, to maintain consistency of definition, description and visualization.
Construction of the splicing graphs
Gene structure information including the location of the transcript and the start and end positions of each exon/intron that make up the transcript model are checked for consistency and then loaded into a list. All transcripts are converted to the leading strand for consistency. The exon list is then retrieved and clustered on the basis of the exons occupying overlapping genomic positions. When exons overlap, the server checks for well-determined borders and also which exons occur in the majority of transcripts and retains these as distinct exons. If an exon is completely contained in another larger exon, these are not merged but retained as individual exons and then entered into a list maintaining the mapping of variable exons to distinct exons. Splicing graphs are then drawn using these clusters of transcripts from the node list. The first line of the resultant splicing graph is composed entirely of distinct reference exons, followed by subsequent lines showing the locations of variable exons. The exons are connected by edges, representing introns in the set of transcripts provided. Since the splicing graph could be interpreted as paths corresponding to hypothetical transcripts, the observed or input transcript set is also shown, indicating true paths through the graph.
Classification of alternative splicing
ASGS follows the classification schema described in DEDB (8). Rules are derived to detect specific AS events from the distinct and variable exon sets (details and examples of the rules are available on the website). Different from ASG (7), we have an enlarged set of AS classification events. Apart from the classical AS events like cassette exons, intron-retention, alternative donor sites and alternative acceptor sites, we have also elected to classify other gene structure events like alternative transcriptional start/termination sites as well as alternative initiation/termination exons (8). The reason for the existence of the alternative initiation/termination exon categories is due to the fact that the 5′ and 3′ ends of the transcripts are usually difficult to determine experimentally and are thus less accurate. Therefore, any differences in the start and end positions of the transcripts could be simply due to the sequencing difficulties. The inclusion of the alternative initiation/termination exons category is an attempt to circumvent this problem as alternative initiation/termination exons (which are classified based on the end position of initiation exons and the start position of termination exons) are unaffected by the sequencing difficulties and thus represent true alternative exons. Alternative transcriptional start/termination sites, however, are dependent on sequencing results and provide a means of classifying gene segments with differences in the start positions of initiation exons and the end positions of termination exons, with a view to update entries in this category, when the 5′ and 3′ ends of these transcripts are determined accurately. These rules are applied to each splicing graph generated by the server.
SAMPLE OUTPUT
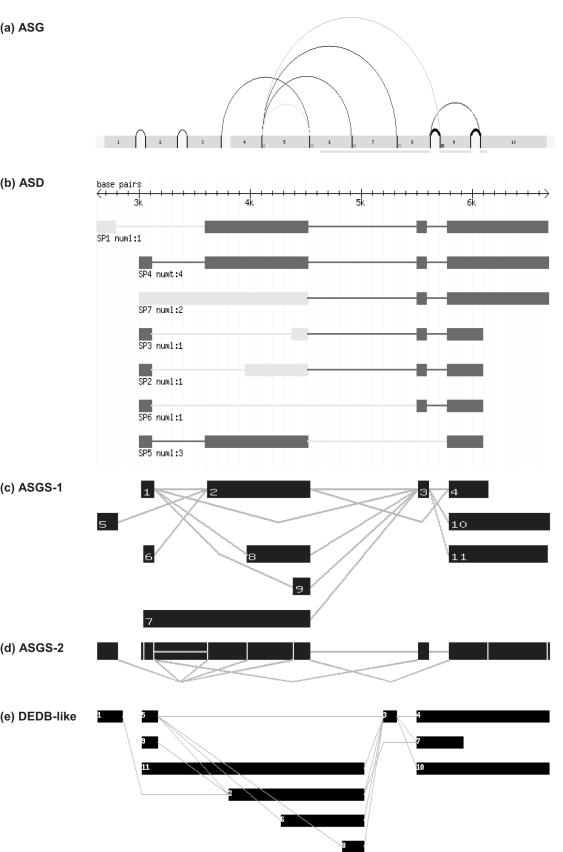
An example of the splicing graph and single line diagram for a human gene [HYAL1 or hyaluronoglucosaminidase 1, ENSG00000114378 (10)] with seven transcripts is shown in Figure 1. We have provided the transcript summary images from ASG (7) and Alternate Splice Database (ASD) (10) for comparison purposes, to highlight the utility of the splicing graph approach over the conventional representation of each transcript as separate lines, used in the ASD database.
Figure 1.
Transcript diversity information for the human hyaluronidase gene, HYAL1 (hyaluronoglucosaminidase 1, ENSG00000114378) (a) from the ASG (7), (b) from the ASD (10) and from ASGS (c) as a splicing graph and (d) a single line splice diagram and (e) the splicing graph diagram using the DEDB formalism (8).
The ASG graphic (Figure 1a) is comprehensive in its presentation of transcripts and AS events are colour-coded. The ASD representation of the same gene (Figure 1b) is a set of individual transcripts. The ASGS transcript splicing graph (Figure 1c) provides a compact representation of all transcripts, with exons organized into distinct reference and variable nodes. The distinct exons form the first line of the splicing graph, with other exons arranged in subsequent lines. The nodes are connected by edges representing the introns in the transcripts shown in Figure 1b. The condensed single line splicing graph (Figure 1d) permits the identification of all splicing junctions. For comparison with our earlier approach, adopted in DEDB (8), where the first transcript was taken as the reference sequence, we note that our present approach is able to identify four reference exons that occur in five, three, six and four of the seven transcripts, respectively. This would be value in comparing AS events occurring in the same gene in different species or under different environmental and disease conditions.
SPLICING GRAPHS FOR HUMAN GENES
AS has emerged as a major causative agent for several human diseases (5). To facilitate the creation of splicing graphs for human genes of interest, we have added an interactive splicing graph generation utility, iASGS, which can read transcript data directly from the curated ASD (10), without generating an input GFF file. ASD provides a user-friendly search facility, for locating specific alternatively spliced human genes. The EnsEMBL identifier for each selected human gene is then provided to iASGS for dynamic retrieval of transcript information from ASD, followed by processing by ASGS. As the transcript information for human genes is not stored permanently on the ASGS server, the splicing graph images and analyses will be generated based on the primary data updated and provided by ASD. This eliminates the need to maintain the data locally and update it periodically to mirror the ASD content. The ASGS output provides compact and single line splicing graph images and tables with exon and AS event information. This utility can also be used for alternatively spliced mouse genes, provided by ASD.
FUTURE WORK
Following this approach, ASGS will focus on developing XML standards for comparing AS events in different genomes by representing them as splicing graphs.
Acknowledgments
D.P. is grateful to the Macquarie University for the award of a research scholarship (iMURS) and a travel grant (MUPGR), facilitating useful discussions with Dr T. A. Thanaraj and the EBI ASD team. Funding to pay the Open Access publication charges for this article was provided by Macquarie University.
Conflict of interest statement. None declared.
REFERENCES
- 1.Sharp P.A. Split genes and RNA splicing. Cell. 1994;77:805–815. doi: 10.1016/0092-8674(94)90130-9. [DOI] [PubMed] [Google Scholar]
- 2.Gilbert W. Why genes in pieces? Nature. 1978;271:501. doi: 10.1038/271501a0. [DOI] [PubMed] [Google Scholar]
- 3.International Human Genome Sequencing Consortium. Finishing the euchromatic sequence of the human genome. Nature. 2004;431:931–945. doi: 10.1038/nature03001. [DOI] [PubMed] [Google Scholar]
- 4.Stetefeld J., Ruegg M.A. Structural and functional diversity generated by alternative mRNA splicing. Trends Biochem. Sci. 2005;30:515–521. doi: 10.1016/j.tibs.2005.07.001. [DOI] [PubMed] [Google Scholar]
- 5.Hagiwara M. Alternative splicing: a new drug target of the post-genome era. Biochim. Biophys. Acta. 2005;1754:324–331. doi: 10.1016/j.bbapap.2005.09.010. [DOI] [PubMed] [Google Scholar]
- 6.Lee C., Atanelov L., Modrek B., Xing Y. ASAP: the Alternative Splicing Annotation Project. Nucleic Acids Res. 2003;31:101–105. doi: 10.1093/nar/gkg029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Leipzig J., Pevzner P., Heber S. The Alternative Splicing Gallery (ASG): bridging the gap between genome and transcriptome. Nucleic Acids Res. 2004;32:3977–3983. doi: 10.1093/nar/gkh731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lee B.T., Tan T.W., Ranganathan S. DEDB: a database of Drosophila melanogaster exons in splicing graph form. BMC Bioinformatics. 2004;5:189. doi: 10.1186/1471-2105-5-189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kim P., Kim N., Lee Y., Kim B., Shin Y., Lee S. ECgene: genome annotation for alternative splicing. Nucleic Acids Res. 2005;33:D75–D79. doi: 10.1093/nar/gki118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stamm S., Riethoven J.J., Le Texier V., Gopalakrishnan C., Kumanduri V., Tang Y., Barbosa-Morais N.L., Thanaraj T.A. ASD: a bioinformatics resource on alternative splicing. Nucleic Acids Res. 2006;34:D46–D55. doi: 10.1093/nar/gkj031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Holste D., Huo G., Tung V., Burge C.B. HOLLYWOOD: a comparative relational database of alternative splicing. Nucleic Acids Res. 2006;34:D56–D62. doi: 10.1093/nar/gkj048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee C., Wang Q. Bioinformatics analysis of alternative splicing. Brief. Bioinform. 2005;6:23–33. doi: 10.1093/bib/6.1.23. [DOI] [PubMed] [Google Scholar]
- 13.Modrek B., Lee C. A genomic view of alternative splicing. Nature Genet. 2002;30:13–19. doi: 10.1038/ng0102-13. [DOI] [PubMed] [Google Scholar]
- 14.Heber S., Alekseyev M., Sze S.H., Tang H., Pevzner P.A. Splicing graphs and EST assembly problem. Bioinformatics. 2002;18:S181–S188. doi: 10.1093/bioinformatics/18.suppl_1.s181. [DOI] [PubMed] [Google Scholar]
- 15.Eyras E., Reymond A., Castelo R., Bye J.M., Camara F., Flicek P., Huckle E.J., Parra G., Shteynberg D.D., Wyss C., et al. Gene finding in the chicken genome. BMC Bioinformatics. 2005;6:131. doi: 10.1186/1471-2105-6-131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. GFF: an Exchange Format for Feature Description. http://www.sanger.ac.uk/Software/formats/GFF/, last accessed on 13/02/2006.
- 17.Bollina D., Lee B.T.K., Ranganathan S. Kotsis G., Tanier D., Bressan S., Ibrahim I.K., Mokhtar S. MVC Architecture in Bioinformatics web applications and its Java implementation. The seventh International Conference on Information Integration and Web-based Applications and Services (iiWAS2005); September 19–21; Kuala Lumpur, Malaysia. 2005. pp. 759–764. Austrian Computer Society, vol. 2. [Google Scholar]