Abstract
The internal transcribed spacer 2 (ITS2) is a phylogenetic marker which has been of broad use in generic and infrageneric level classifications, as its sequence evolves comparably fast. Only recently, it became clear, that the ITS2 might be useful even for higher level systematic analyses. As the secondary structure is highly conserved within all eukaryotes it serves as a valuable template for the construction of highly reliable sequence-structure alignments, which build a fundament for subsequent analyses. Thus, any phylogenetic study using ITS2 has to consider both sequence and structure. We have integrated a homology based RNA structure prediction algorithm into a web server, which allows the detection and secondary structure prediction for ITS2 in any given sequence. Furthermore, the resource contains more than 25 000 pre-calculated secondary structures for the currently known ITS2 sequences. These can be taxonomically searched and browsed. Thus, our resource could become a starting point for ITS2-based phylogenetic analyses and is therefore complementary to databases of other phylogenetic markers, which focus on higher level analyses. The current version of the ITS2 database can be accessed via http://its2.bioapps.biozentrum.uni-wuerzburg.de.
INTRODUCTION
The internal transcribed spacer 2 (ITS2) is part of the eukaryotic nuclear rDNA cistron and lies between the 5.8S and the 28S rRNA. A remarkable feature of this sequence is its high divergence between species. Thus it is a well suited marker for low level phylogenetic analyses. Indeed, after its first application in 1991 (1), it has been used on a rapidly growing number of publications, totalling to 1218 publications concerning the ITS2. But, the high divergence of the sequence, although enabling low level classifications is a hindrance for the application of this marker for more general phylogenetic analyses.
However, when comparing the structure of this RNA molecule, it turned out that a conserved core can be found between species as divergent as vertebrates and yeast (2) or green algae and higher plants (3). This core consists of four helices with the third as the longest. Additionally, two motifs have been suggested (4). If this structural core is indeed a general feature of the ITS2, it could complement the highly divergent sequence with a more slowly evolving feature. Accordingly, the combination of both might enable the application of the ITS2 for low and higher level analyses (4). Indeed, we could show recently, that the secondary structure core can be found within a wide-range of eukaryotes (5), resulting in more than 5000 ITS2 sequences and their associated secondary structures. This analysis also revealed, that the core structure is found only rarely by standard RNA folding programs. We therefore developed a method based on homology modelling enabling the prediction of the secondary structure for a wider range of ITS2. Indeed, basing on the 5000 structures predicted by standard RNA folding programs, we could identify an additional 20 000 ITS2 structures following the conserved core (6). Here, we describe (i) a web server for the homology modelling of novel sequences and (ii) the web-based access to 25 000 ITS2 sequences and their individual secondary structures.
STRUCTURE PREDICTION
We have recently developed and applied a method for the homology based prediction of the ITS2 secondary structure. It was based on the Needleman–Wunsch algorithm (7) leading to global optimal alignments between a sequence with a known structure and a novel sequence. Although suitable for a standalone application, this approach contains several hindrances for a web-based interface. First, as global alignments are needed for the assignment of the whole structure, the boundaries of the unknown ITS2 have to be known. Second, global alignments between a user supplied sequence and the database of 25 000 sequences will be too time consuming. We therefore modified the original approach by performing a BLAST (8) search with the novel sequence against the database of sequences with known structures to (i) estimate the position of the ITS2 within the sequence and (ii) identify candidates for homology modelling. In a second step, models using all candidates are calculated. Only those models fulfilling defined quality criteria are reported.
Within the BLAST search, we are using an ITS2 specific score matrix estimated from a large set of reliable ITS2 sequence alignments (9). As the matrix deviates substantially from the default BLAST nucleotide score matrix, results of database searches as well as the revealed alignments will be more accurate than those estimated in standard searches. All hits with an E-value <10−10 are considered for further evaluation. As Blast delivers local alignments, the positions of the boundaries of the ITS2 in the unknown sequence have to be estimated. Therefore, the length of the hit within the novel sequence is extended in both directions according to the length of the template. Finally, the structure of the novel ITS2 is homology modelled using each hit as template according to the method described in Wolf et al. (6). To assure reliable models, only those, where at least 75% of the structure for each helix could be transferred, are reported. This is in accordance with the criteria applied in the original publication of the homology modelling approach. As shown in Figure 1, the predicted structure together with the alignment to the template sequence is displayed as a 2D model as well as in bracket notation.
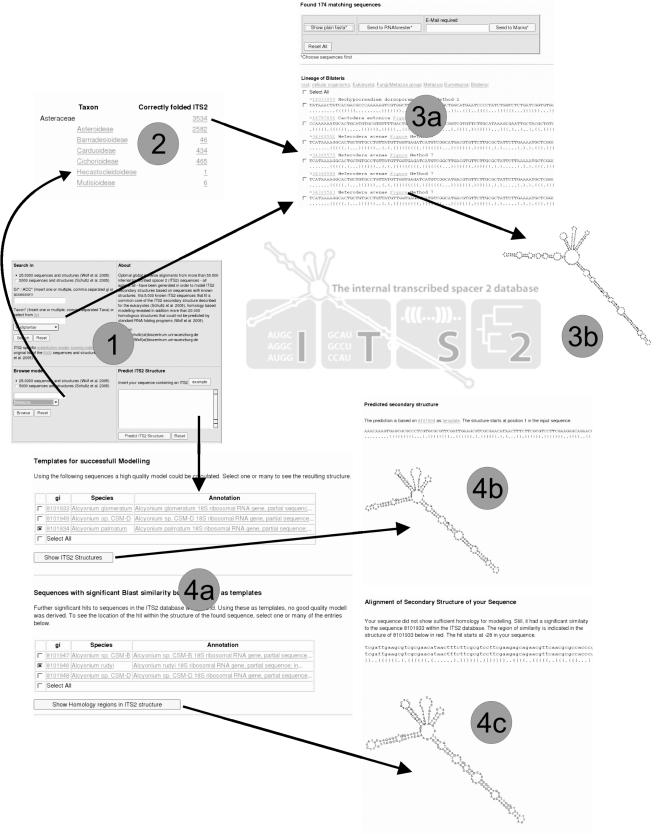
Figure 1.
Screen shots of the web interface—(1) Start and Input view; (2) results of browsing for Asteraceae; (3a) list of ITS2 sequences and secondary structures in bracket notation for Bilateria, (3b) a sample secondary structure as stored in the database; (4) modelling of an ITS2 structure (4a) results of the BLAST search leading to high and low quality models, (4b) a predicted high quality model, (4c) homologous region indicated on the secondary structure of the template.
In cases where no high quality models can be calculated despite a significant Blast hit, the position of the similarity within the template structure is displayed (Figure 1). As multiple hits can be presented at the same time, one can easily get an overview of the consistency of the homology region.
SEARCHING AND BROWSING
The prediction of the structure of a novel ITS2 will in most cases be the first step in a phylogenetic analysis. In a second step, the novel sequence has to be complemented with further sequences of the taxon of interest. Again, those with a known structure might be of highest use. We therefore implemented the ability to search for ITS2 sequences with correctly predicted structures (according to our model) not only within a species but within each taxon of interest. As a taxonomic classification, we are using the NCBI taxonomy database (with all its limitations according to phylogeny). In many cases, a taxonomic rank for the phylogenetic classification might not be known beforehand but depend on the coverage of the marker within the taxon. To provide a first overview, we implemented a browse mode, which allows traversing the NCBIs taxonomy (Figure 1). For each node and its siblings the number of ITS2 with predicted structures is given. Once the level of interest is reached, the ITS2 sequence and structure for all species belonging to this node can be retrieved.
The resulting list of sequence-structure pairs can be the starting point for further analyses. To enable the upload to other, third party programs, the list can be exported in fasta format. A very common application will be the alignment of two or more sequences based on sequence and structure, in order to perform a phylogenetic analysis. Currently, there are only a few programs calculating global RNA alignments including sequence and structure information. Starting from the sequence-structure pairs of the ITS2 database, we allow to directly calling two of these programs, namely MARNA (10) and RNAforester (11) with selected sequences.
CONCLUSION
Already the ITS2 sequences combined with their secondary structures stored in the database might be of substantial interest for phylogenetic analyses. For example a recent analysis of the family Asteraceae (the daisy family) relied on 340 ITS2 sequences (12). To date, there are 3534 ITS2 sequences for this family with correctly folded structures within our database. A re-examination of this family considering these data might give a substantially broader coverage and thereby deeper insights into the evolution of this taxon. The collection of ITS2 together with the ability to predict the structure of novel sequences makes the ITS2 database a starting point for detailed phylogenetic analyses. Therefore, it is of importance to make the interaction with further tools for phylogenetic analyses straightforward. On the one hand, we have already integrated access to MARNA (10) and RNAforester (11), enabling the structure and sequence based alignment of ITS2 sequences. On the other hand, we provide a fasta formatted output which can be used as input for further programs. Once an alignment is calculated, standard phylogenetic programs like Phylip or more specific ones like the CBCanalyzer (13) can be used for further analyses. In the near future, the ability to calculate sequence and structure based alignments as well as to edit them will be added to the web server (Seibel, P.N., Müller, T., Dandekar, T., Schultz, J. and Wolf, M., manuscript in preparation). Finally, to enable the integration of our resource into third party projects, we provide a SOAP based interface (for details see Web page).
In summary, the described resource can be a starting point for any ITS2-based phylogenetic analysis and thereby complement databases for other phylogenetic markers like the European ribosomal database (14). Whereas the first might be of special use for low level analyses focussing on species and genus, the second might be more suited for higher level analyses. The combination of both will therefore give increased insight over a wide-range of taxonomic levels.
Acknowledgments
Funding to pay the Open Access publication charges for this article was provided by Helmholtz Society (VH-VI-023).
Conflict of interest statement. None declared.
REFERENCES
- 1.Porter C.H., Collins F.H. Species-diagnostic differences in a ribosomal DNA internal transcribed spacer from the sibling species Anopheles freeborni and Anopheles hermsi (Diptera:Culicidae) Am. J. Trop. Med. Hyg. 1991;45:271–279. doi: 10.4269/ajtmh.1991.45.271. [DOI] [PubMed] [Google Scholar]
- 2.Joseph N., Krauskopf E., Vera M.I., Michot B. Ribosomal internal transcribed spacer 2 (ITS2) exhibits a common core of secondary structure in vertebrates and yeast. Nucleic Acids Res. 1999;27:4533–4540. doi: 10.1093/nar/27.23.4533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mai J.C., Coleman A.W. The internal transcribed spacer 2 exhibits a common secondary structure in green algae and flowering plants. J. Mol. Evol. 1997;44:258–271. doi: 10.1007/pl00006143. [DOI] [PubMed] [Google Scholar]
- 4.Coleman A.W. ITS2 is a double-edged tool for eukaryote evolutionary comparisons. Trends Genet. 2003;19:370–375. doi: 10.1016/S0168-9525(03)00118-5. [DOI] [PubMed] [Google Scholar]
- 5.Schultz J., Maisel S., Gerlach D., Müller T., Wolf M. A common core of secondary structure of the internal transcribed spacer 2 (ITS2) throughout the Eukaryota. RNA. 2005;11:361–364. doi: 10.1261/rna.7204505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wolf M., Achtziger M., Schultz J., Dandekar T., Müller T. Homology modeling revealed more than 20 000 rRNA internal transcribed spacer 2 (ITS2) secondary structures. RNA. 2005;11:1616–1623. doi: 10.1261/rna.2144205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Needleman S.B., Wunsch C.D. A general method applicable to the search for similarities in the amino acid sequence of two proteins. J. Mol. Biol. 1970;48:443–453. doi: 10.1016/0022-2836(70)90057-4. [DOI] [PubMed] [Google Scholar]
- 8.Altschul S.F., Madden T.L., Schaffer A.A., Zhang J., Zhang Z., Miller W., Lipman D.J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Muller T., Vingron M. Modeling amino acid replacement. J. Comput. Biol. 2000;7:761–776. doi: 10.1089/10665270050514918. [DOI] [PubMed] [Google Scholar]
- 10.Siebert S., Backofen R. MARNA: multiple alignment and consensus structure prediction of RNAs based on sequence structure comparisons. Bioinformatics. 2005;21:3352–3359. doi: 10.1093/bioinformatics/bti550. [DOI] [PubMed] [Google Scholar]
- 11.Höchsmann M., Töller T., Giegerich R., Kurtz S. Local similarity in RNA secondary structures. Proceedings of the IEEE Bioinformatics Conference 2003; 2003. pp. 159–168. [PubMed] [Google Scholar]
- 12.Goertzen L.R., Cannone J.J., Gutell R.R., Jansen R.K. ITS secondary structure derived from comparative analysis: implications for sequence alignment and phylogeny of the Asteraceae. Mol. Phylogenet. Evol. 2003;29:216–234. doi: 10.1016/s1055-7903(03)00094-0. [DOI] [PubMed] [Google Scholar]
- 13.Wolf M., Friedrich J., Dandekar T., Muller T. CBCAnalyzer: inferring phylogenies based on compensatory base changes in RNA secondary structures. In Silico Biol. 2005;5:291–294. [PubMed] [Google Scholar]
- 14.Wuyts J., Perriere G., Van De Peer Y. The European ribosomal RNA database. Nucleic Acids Res. 2004;32:D101–D103. doi: 10.1093/nar/gkh065. [DOI] [PMC free article] [PubMed] [Google Scholar]