Abstract
MEME (Multiple EM for Motif Elicitation) is one of the most widely used tools for searching for novel ‘signals’ in sets of biological sequences. Applications include the discovery of new transcription factor binding sites and protein domains. MEME works by searching for repeated, ungapped sequence patterns that occur in the DNA or protein sequences provided by the user. Users can perform MEME searches via the web server hosted by the National Biomedical Computation Resource (http://meme.nbcr.net) and several mirror sites. Through the same web server, users can also access the Motif Alignment and Search Tool to search sequence databases for matches to motifs encoded in several popular formats. By clicking on buttons in the MEME output, users can compare the motifs discovered in their input sequences with databases of known motifs, search sequence databases for matches to the motifs and display the motifs in various formats. This article describes the freely accessible web server and its architecture, and discusses ways to use MEME effectively to find new sequence patterns in biological sequences and analyze their significance.
INTRODUCTION
The purpose of MEME (Multiple EM For Motif Elicitation) (rhymes with ‘team’) (1,2) is to allow users to discover signals (called ‘motifs’) in DNA or protein sequences. The user of MEME inputs a set of sequences believed to share some (unknown) sequence signal(s). For example, some or all of a set of promoters from co-expressed and/or orthologous genes may contain binding sites (the ‘signal’) for the same transcription factor (3). Similarly, a set of proteins that interact with a single host protein may do so via similar domains (the ‘signal’) (4). Both types of sequence signals can often be represented as motifs-ungapped, approximate sequence patterns. Using a process akin to gapless, local, multiple sequence alignment, MEME searches for statistically significant motifs in the input sequence set. In this way, MEME can discover the binding sites for the shared transcription factor in the set of promoters or the common protein–protein binding domains in the set of proteins. MEME can also be used to discover motifs describing many other types of DNA or protein signals besides transcription factor binding sites and protein–protein interaction domains.
To use MEME via the website, the user provides a set of sequences in the FASTA format by either uploading a file or by cut-and-paste. The only other required input is an email address where the results will be sent. (A planned future version will remove this requirement by providing temporary storage of the results on the web server for a preset period of time.) By default, MEME looks for up to three motifs, each of which may be present in some or all of the input sequences. MEME chooses the width and number of occurrences of each motif automatically in order to minimize the ‘E-value’ of the motif—the probability of finding an equally well-conserved pattern in random sequences. By default, only motif widths between 6 and 50 are considered, but the user may change this as well as several other aspects of the search for motifs.
The MEME output is HTML and shows the motifs as local multiple alignments of (subsets of) the input sequences, as well as in several other formats (Figure 1). ‘Block diagrams’ show the relative positions of the motifs in each of the input sequences. Buttons on the MEME HTML output allow one or all of the motifs to be forwarded for analysis by other web-based programs. Clicking on a button allows all of the motifs to be sent to the MAST web server where various sequence databases (or uploaded sequences) can be searched for sequences matching the motifs. This is useful in cases, for example, where the user would like to find whether the motif of interest is also present in other genes or genomes.
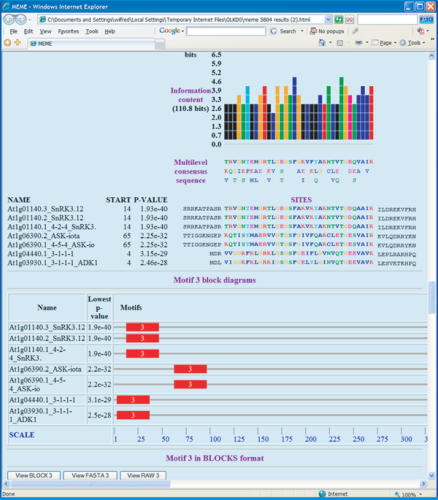
Figure 1.
Sample MEME output.This portion of an MEME HTML output form shows a protein motif that MEME has discovered in the input sequences. The sites identified as belonging to the motif are indicated, and above them is the ‘consensus’ of the motif and a color-coded bar graph showing the conservation of each position in the motif. Some of the hyperlinked buttons that allow the motif to be viewed and analyzed in other ways can be seen at the bottom of the screen shot.
MAST is a web-based tool that can be used to search for sequences that match one or more motifs. It can be used to look for sequences that contain motifs found by MEME, by other motif discovery tools or that are taken from a motif database. The MAST website, reached via the same URL as the MEME website, provides numerous nucleotide and protein databases for searching. MAST queries may contain any number of motifs, and it scores each sequence in the selected database using all of the motifs. In the first example above, MAST can search DNA sequences for matches to the putative transcription factor binding site (TFBS) motifs found by MEME in a set of promoter sequences. MAST can search for matches in protein sequences to the putative protein–protein interaction motifs found in the second MEME example.
Users of MEME via the website or locally installed versions are asked to cite this article as well as the primary reference for MEME (5). Users of MAST are asked to cite this article and Ref. (6).
MOTIF DISCOVERY STRATEGIES
Motif discovery can be viewed as a ‘needle in a haystack’ problem. The motif discovery algorithm is looking for a set of similar short sequences (the needle) in a set of much longer sequences (the haystack). The problem is easier when the motif instances are long and very similar to each other. It gets much harder when the motif instances are short and/or degenerate, or the input sequences are very long.
Discovering TFBS motifs in a set of DNA sequences (e.g. genomic regions upstream of genes) is a difficult task owing to the tendency of binding sites to be short and degenerate, and owing to the fact that promoter regions are often difficult to identify precisely. The problem tends to be worse in eukaryotes than in prokaryotes and yeast because eukaryotic TFBS tend to be shorter and more variable (7).
To successfully discover TFBS motifs with MEME, it is necessary to choose and prepare the input sequences carefully. Candidate sequences can be the promoters of genes believed to be co-regulated based on the evidence from expression microarray experiments, or sequences appearing to bind to a transcription factor based on chromatin immunoprecipitation experiments. The sequences should be as short as possible and contain as few ‘noise’ sequences (sequences not containing any motif) as possible. Ideally, the sequences should be <1000 bp long (8). Including more than 40 motif-containing sequences generally does not improve TFBS motif discovery with MEME and similar algorithms (9). If the sequences contain low-information segments that do not contain motifs of interest, it can be helpful to remove them using the DUST program (R. L. Tatusov and D. J. Lipman, unpublished NCBI/Toolkit), which is available for downloading at http://blast.wustl.edu/pub/dust/. Repetitive DNA elements should also be removed from the sequences input to MEME using the RepeatMasker program (A. Smit, R. Hubley and P. Green, unpublished data), which can be accessed via the Web (http://www.repeatmasker.org/cgi-bin/WEBRepeatMasker).
It should be noted that MEME is not suited to whole-genome TFBS motif discovery. Owing to their shortness and degeneracy, TFBS motifs become statistically ‘invisible’ in the context of a whole genome. The sensitivity of the search for TFBS motifs can be improved by using a ‘higher-order background sequence model’, but this option is only available currently when users download the MEME source code and install it locally. Instructions for the installation are available at the MEME website (http://meme.nbcr.net/meme/website/meme-download.html) by clicking on ‘View MEME man page’; see the documentation for the ‘-bfile’ switch there.
Protein motifs are generally easier to discover owing to the length of the protein alphabet and the chemical similarity among groups of amino acids. This allows shorter motifs to be more statistically significant and makes it easier to distinguish functional motifs from statistical artifacts. To use MEME to discover protein motifs, the same basic guidelines apply as with DNA motifs—keep the sequences as short as possible and include as few sequences that are not likely to contain the motif as possible in the input to MEME. Low-complexity regions can be removed from the protein input sequences using the SEG program (10).
ANALYZING MOTIFS USING THE MEME OUTPUT HYPERLINKS
The MEME HTML output contains buttons making it easy to analyze the motifs it discovers. By clicking on the button labeled ‘Compare PSPM to known motifs in JASPAR database’ following each motif, the DNA motif can be compared to each of the motifs in the JASPAR database (11) of known TFBS motifs. Similarly, protein motifs may be compared with protein motifs in the BLOCKS database of protein motifs (12) by clicking on the ‘submit BLOCK’ button following each motif on the MEME form. This takes the user to the ‘BLOCKS server’ where clicking on ‘LAMA’ will compare the motif with those in the BLOCKS database. The BLOCKS server also allows users to display protein motifs in many different ways, including LOGOS (13) or phylogenetic trees, by clicking on the corresponding buttons on the BLOCKS server form. By clicking on one of the file output formats under Logos, the user is able to obtain a LOGOS diagram similar to that shown in Figure 2.
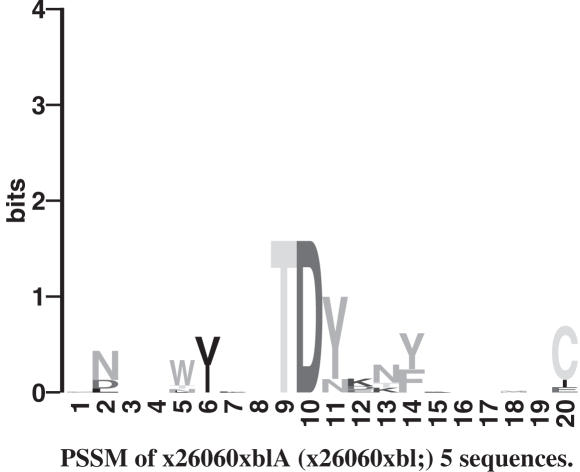
Figure 2.
LOGO of protein motif. LOGOS are a visualization tool for motifs. The height of a letter indicates its relative frequency at the given position (x-axis) in the motif.
To search sequences for matches to the motifs found by MEME, users can click on the ‘MAST’ button at the top of the MEME output form. This will take the user to the MAST website where they can select the database to search. Since MAST is sequence-oriented, TFBS motifs should only be used to search promoter regions. These are listed in the MAST database pull-down menu as ‘Upstream Sequence Databases’. Currently, only a few organisms are supported. However, users can upload their own database of promoter sequences for searching using MAST. Protein motifs can be used to search any of the sequence databases provided by the MAST website since MAST can search either protein or nucleotide databases with protein motifs. The MAST database are updated weekly.
WEB SERVER AND USER SUPPORT
As of MEME version 3.5, the configuration and installation of MEME (including the web server) is significantly simplified by using Autoconf (http://www.gnu.org/software/autoconf/autoconf.html) and Automake (http://www.gnu.org/software/automake/automake.html) from the GNU Build System. An installation session for MEME and MAST web server may be as simple as follows:
cd meme_3.5.2
./configure --prefix=$HOME/meme --with-url=http://www.nbcr.net/
meme --enable-web
make
make test
make install
Supported platforms now include Linux, Solaris, MacOS X, Cygwin and Irix.
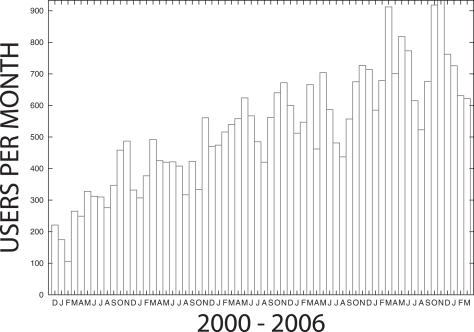
The MEME web server hosted by NBCR is queried by about 800 different users (based on unique email addresses) each month. Usage has been growing steadily since the service was first introduced in 1996. Figure 3 shows usage growth at the NBCR server since 2000.
Figure 3.
Usage of MEME at the NBCR web server. The plot shows the number of different users submitting jobs to the NBCR MEME web server each month since December 2000. Usage figures for March 2006 include up to March 20 only.
To meet the growing user demand and take advantage of the emerging grid-computing resources (14), we have made MEME available for the installation on Linux clusters using either the RPM package manager or Rocks. The RPM package manager is a tool for managing software installation on computers running many versions of the Linux operating system. Rocks (http://www.rocksclusters.org) is a highly customized toolkit for computational biologists and engineers to build and maintain Linux clusters. The current NBCR MEME web server cluster is built using the MEME roll for Rocks and requires minimal maintenance effort.
MEME and MAST can be downloaded and installed free of charge by academic users via the website: (http://meme.nbcr.net/meme/website/meme-download.html). Approximately 300 users download the MEME/MAST software each month. The MEME support team offers assistance to the MEME and MAST user community through the forum (http://nbcr.net/forum/viewforum.php?f=5) or the mailing list (meme@nbcr.net). Institutes interested in setting up MEME mirror sites are encouraged to contact us for any assistance.
FUTURE DIRECTIONS
To increase the sensitivity of MEME searches, we will add an option in the web server to let the user upload a background sequence model to MEME. We hope to add algorithms for removing low-complexity regions (SEG and DUST) and repeated elements (RepeatMasker) in the MEME website as a convenience to users. These services will also be exposed as web services and are integrated using workflow tools developed by using NBCR.
We have also planned to add buttons to the MEME output to allow TFBS motifs to be used in searching for cis-regulatory modules via algorithms such as MCAST (15). MCAST will be configured to be able to search the same DNA databases as MAST. In conjunction with this, we will add databases of upstream sequences for many additional organisms to the MAST/MCAST websites to facilitate the analysis of TFBS motifs discovered by using MEME.
NBCR has developed a set of tools built on top of the open source software that allows bioinformatics applications to be deployed as Web Services easily (S. Krishnan, B. Stearn, K. Bhatia, W. W. Li and P. Arzberger, manuscript submitted) and leverage the Cyberinfrastructure components transparently (14). A prototype has been deployed using MEME as a scientific driver (16) that offers a user with a dynamic pool of distributed compute resource, workflow management console and a friendly user interface. This portal will be deployed to the production web server in the future.
Acknowledgments
The authors acknowledge NBCR award from NCRR, NIH P41 RR08605, for support of the MEME and MAST website. TLB acknowledges grant from NIH, R01 RR021692-01, for support of continuing development of the MEME and related sequence analysis tools. T.L.B. also acknowledges the ARC Centre for Bioinformatics (ACB) (ARC CE0348221) for infrastructure support for the MEME mirror site at the ACB. Funding to pay the Open Access publication charges for this article was provided by the NIH.
Conflict of interest statement. None declared.
REFERENCES
- 1.Bailey T.L., Elkan C. Unsupervised Learning of Multiple Motifs In Biopolymers Using EM. Mach. Learn. 1995;21:51–80. [Google Scholar]
- 2.Bailey T.L., Elkan C. The value of prior knowledge in discovering motifs with MEME. In: Rawlings C., Clark D., Altman R., Hunter L., Lengauer T., Wodak S., editors. Proceedings of the Third International Conference on Intelligent Systems for Molecular biology, July; Menlo Park, CA: AAAI Press; 1995. pp. 21–29. [PubMed] [Google Scholar]
- 3.Lyons T.J., Gasch A.P., Alex Gaither L., Botstein D., Brown P.O., Eide D.J. Genome-wide characterization of the Zap1p zinc-responsive regulon in yeast. Proc. Natl Acad. Sci. USA. 2000;97:7957–7962. doi: 10.1073/pnas.97.14.7957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fang J., Haasl R.J., Dong Y., Lushington G.H. Discover protein sequence signatures from protein-protein interaction data. BMC Bioinformatics. 2005;6:1–8. doi: 10.1186/1471-2105-6-277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bailey T.L., Elkan C. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. In: Altman R.B., Brutlag D.L., Karp P.D., Lathrop R.H., Searls D.B., editors. Proceedings of the Second International Conference on Intelligent Systems for Molecular Biology, August; Menlo Park, CA: AAAI Press; 1994. pp. 28–36. [PubMed] [Google Scholar]
- 6.Bailey T.L., Gribskov M. 'Combining evidence using P-values: application to sequence homology searches. Bioinformatics. 1998;14:48–54. doi: 10.1093/bioinformatics/14.1.48. [DOI] [PubMed] [Google Scholar]
- 7.Tompa M., Li N., Bailey T.L., Church G.M., De Moor B., Eskin E., Favorov A.V., Frith M.C., Fu Y., Kent W.J., et al. Assessing Computational Tools for the Discovery of Transcription Factor Binding Sites. Nat. Biotechnol. 2005;23:137–147. doi: 10.1038/nbt1053. [DOI] [PubMed] [Google Scholar]
- 8.Pevzner P.A., Sze S.H. Combinatorial approaches to finding subtle signals in DNA sequences. In: Bourne P.E., Gribskov M., Altman R.B., Jensen N., Hope D., Lengauer T., Mitchell J.C., Scheeff E.D., Smith C., Strande S., Weissig H., editors. Proceedings of the Eighth International Conference on Intelligent Systems for Molecular Biology, August.; Menlo Park, CA: AAAI Press; 2000. pp. 269–278. [PubMed] [Google Scholar]
- 9.Hu J., Li B., Kihara D. Limitations and potentials of current motif discovery algorithms. Nucleic Acids Res. 2005;33:4899–4913. doi: 10.1093/nar/gki791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wootton J.C., Federhen S. Analysis of compositionally biased regions in sequence databases. Methods Enzymol. 1966;266:554–571. doi: 10.1016/s0076-6879(96)66035-2. [DOI] [PubMed] [Google Scholar]
- 11.Sandelin A., Alkema W., Engström P., Wasserman W.W., Lenhard B. JASPAR: an open-access database for eukaryotic transcription factor binding profiles. Nucleic Acids Res. 2004;32:D91–D94. doi: 10.1093/nar/gkh012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Henikoff J.G., Pietrokovski S., Henikoff S. Recent enhancements to the blocks database servers. Nucleic Acids Res. 1997;25:222–225. doi: 10.1093/nar/25.1.222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schneider T.D., Stephens R.M. Sequence logos: a new way to display consensus sequences. Nucleic Acids Res. 1990;18:6097–6100. doi: 10.1093/nar/18.20.6097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Foster I., Kesselman C. The Grid 2: Blueprint for a New Computing Infrastructure. 2nd edn. San Francisco, CA: Morgan Kaufmann Publishers, Inc.; 2004. [Google Scholar]
- 15.Bailey T.L., Noble W.S. Searching for statistically significant regulatory modules. Bioinformatics. 2003;19(Suppl 2):II16–II25. doi: 10.1093/bioinformatics/btg1054. [DOI] [PubMed] [Google Scholar]
- 16.Li W.W., Krishnan S., Mueller K., Misleh C., Arzberger P. Building cyberinfrastructure for bioinformatics using service oriented architecture. In: Bu Sung F.L., Abramson D., Cai W., Graupner S., Jin H., Sloot P., editors. Proceedings of the IEEE International Symposium on Cluster Computing and the Grid, May; USA: IEEE Press; 2006. (in press) [Google Scholar]