Abstract
Predicting domains of proteins is an important and challenging problem in computational biology because of its significant role in understanding the complexity of proteomes. Although many template-based prediction servers have been developed, ab initio methods should be designed and further improved to be the complementarity of the template-based methods. In this paper, we present a novel domain prediction system KemaDom by ensembling three kernel machines with the local context information among neighboring amino acids. KemaDom, an alternative ab initio predictor, can achieve high performance in predicting the number of domains in proteins. It is freely accessible at http://www.iipl.fudan.edu.cn/lschen/kemadom.htm and http://www.iipl.fudan.edu.cn/~lschen/kemadom.htm.
INTRODUCTION
Domains are the structural, functional and evolutionary units of proteins. Most multidomain proteins are formed by duplication, divergence and recombination of domains in the history of evolution (1). Thus domains are a key to understand the evolution of proteomes and their complexities. It is therefore of great importance to predict domains in proteins. The importance of this task has been emphasized by the CASP 6 (http://predictioncenter.org/) and the CAFASP 4 (http://www.cs.bgu.ac.il/dfischer/CAFASP4/ and http://www.cs.bgu.ac.il/~dfischer/CAFASP4/) protein structure prediction experiments. However, predicting domains from sequence remains an open problem.
Previous works exhibit great successes in domain prediction. Most of them are online web servers which can be publicly accessed from Internet. All these methods can be classified into two classes: template-based methods (scoring the sequence against domain templates or secondary structure elements) and ab initio methods (non-template methods). The template-based methods include Robetta-Ginzu (2), http://ekhidna.biocenter.helsinki.fi:9801/sqgraph/pairsdb,ADDA (3); http://bioinf.cs.ucl.ac.uk/dompred/DomPredform.html, Dompred-Domssea (4); Dopro (5); http://www.ebi.ac.uk/InterProScan, InterProScan (6); and http://www.bio.ifi.lmu.de/SSEP/, SSEP-Domain (7). And the ab initio methods include http://biozon.org/tools/domain/, Biozon(8); CHOPnet (9); Armadillo (http://armadillo.blueprint.org/; http://www.ics.uci.edu/baldig/dompro.html), DOMpro (10); http://bioinf.cs.ucl.ac.uk/dompred/DomPredform.html, Dompred-DPS (11); http://globplot.embl.de/, Globplot (12); and http://bioinformatics.cribi.unipd.it/cgi-bin/primex_client.cgi, Mateo (13). Additionally, http://meta-dp.cse.buffalo.edu/ Meta-DP (14) is an integrated domain prediction server which ensembles various template-based and ab initio methods with a ‘majority voting’ strategy.
Template-based methods become less effective when a potential domain shares low similarity with the identified domains. Thus, with the availability of domain databases such as CATH (15), SCOP (16) and FSSP-Dali Domain Dictionary (17), the effective ab initio methods using machine learning techniques have been developed (8–10). These methods using different artificial neural networks with various features have made important contributions to this task. Biozon (8) is a hybrid learning system for domain prediction and adopts a feed-forward network using back-propagation algorithm. In this system, the input units consist of sequence termination, correlation, contact profile, class and amino acid entropy, secondary structure, and physio-chemical properties. CHOPnet (9) also uses a three-layer feed-forward neural network but with different features, including secondary structure, solvent accessibility, HSSP conservation weight, the profile of six critical residues {P, H, D, Y, V, C}, secondary structure difference and flexibility of a five-residue segment. These features are proved to be important to the performance of the network. DOMpro (10) applies the 1D-recursive neural network that leverages evolutionary profiles, predicted secondary structure and relative solvent accessibility. It is ranked among the top ab initio domain predictors in the CAFASP 4 evaluation.
Since the most important step of the ab initio methods for domain prediction is to discriminate boundary residues from domain residues, the prediction can be viewed as a two-class classification problem. As to the classifier, support vector machine (SVM), a classical kernel machine, not only is well-founded theoretically, but also has satisfactory abilities of generalization and avoiding over-fitting (18). Encouraged by the successful applications of SVM in computational biology, including remote protein homology detection (19,20), secondary structure prediction (21,22), and the like, we developed a novel predictor, KemaDom abbreviated from ‘kernel machine for domain prediction’, by ensembling three SVM classifiers, KemaSelf, KemaNeiOne and KemaNeiTwo, with different feature subspaces. The SVM classifiers with different feature subspaces improve the diversity of the result. It makes the ensemble work though SVM is a stable classifier and simply ensembling this kind of classifier with same features is not a good choice. The empirical study has shown that KemaDom has good performance in practice for predicting domains in proteins.
MATERIALS AND METHODS
Training and testing data
Liu et al. (9) have curated a dataset from multiple sources and Cheng et al. (10) have curated another dataset from CATH (15) to avoid the data conflict. In this paper, the latter is used to develop and test the algorithm. In this dataset, a total of 354 multi-domain chains and 963 single-domain chains are retrieved. Among these chains, no pair of sequences share sequence similarity above 25% in a global alignment of length 250. The sequences and the information of secondary structure and solvent accessibility can be obtained from Cheng's website (http://contact.ics.uci.edu/download.html).
In the prediction procedure, we focus on discriminating boundary residues from domain residues. Thus, multi-domain chains are used for training and testing, and single-domain chains are only for testing against the model trained by multi-domain chains. Additionally, a blind set from CAFASP 4 is used as the testing set.
Feature extraction
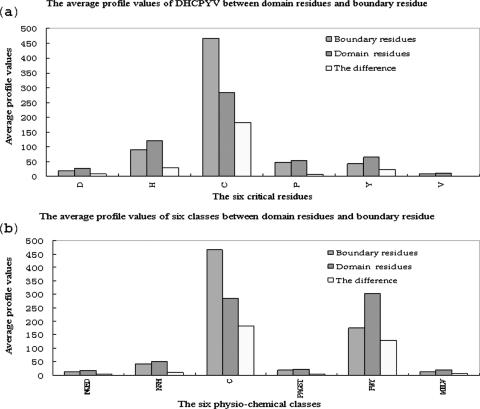
Feature extraction for training and testing is crucial to the model. In our method, we obtain amino acid entropy and physio-chemical properties according to the profile of amino acids. Amino acid entropy measuring the conservation of an alignment can be computed by information entropy. Ferran et al. clustered the 20 residues into 6 classes according to similarity scores of their physio-chemical property (23). One measurement for physio-chemical property is class entropy defined in Ref. (8). Alternatively, we only choose the value of the representative residue from each class to denote physio-chemical property. The six residues are {D, H, C, P, Y, V} because they are most different between domain residues and boundary residues (9). The difference of average profile of critical residues and the difference of average profile of six physio-chemical classes between boundary residues and domain residue (Figure 1) indicate that the latter is more proper as feature units. Secondary structure and relative solvent accessibility can be predicted by widely accepted tools.
Figure 1.
Comparison between average profile of critical residues (a) and the average profile of six physio-chemical classes (b).
According to the above analysis, three sub-models with different input units are designed (Table 1). For KemaSelf, 32 U are extracted as the inputs: 6 U represent physio-chemical information, 1 U represents amino acid entropy, 5 × 3 U are secondary structure of five-residue segment (a center resiude, two left neighborhoods and two right neighborhoods), 5 × 2 U represent solvent accessibility of the segment. For KemaNeiOne (or KemaNeiTwo), 26 U are extracted as the inputs: 2 × 3 U denote secondary structure of the residues with distance d = 1 (or d = 2) from the center residue, 2 × 2 U encode solvent accessibility of those residues, 2 U are amino acid entropy, 2 × 6 U denote physio-chemical properties and the last 2 U allow the exceeding of the N-terminus or C-terminus of the chain.
Table 1.
Features of the sub-models
| Model | Unit position | Description |
|---|---|---|
| KemaSelf | 1–5 | Secondary structure and solvent accessibility of a center residue; |
| 6–11 | Physio-chemical properties of a center residue; | |
| 12–31 | Secondary structure and solvent accessibility of residues with 0 < d ≤ 2; | |
| 32 | Amino acid entropy of a center residue; | |
| KemaNeiOne | 1–6 | Secondary structure of the residues with d = 1; |
| 7–10 | Solvent accessibility of the residues with d = 1; | |
| 11–22 | Physio-chemical properties of the residues with d = 1; | |
| 23–24 | Amino acid entropy of the neighboring residues with d = 1; | |
| 25–26 | Labels to denote the exceeding of the N-terminus or C-terminus of the chain. | |
| KemaNeiTwo | 1–6 | Secondary structure of the left residues with d = 2; |
| 7–10 | Solvent accessibility of the left residues with d = 2; | |
| 11–22 | Physio-chemical properties of the left residues with d = 2; | |
| 23–24 | Amino acid entropy of the neighboring residues with d = 2; | |
| 25–26 | Labels to denote the exceeding of the N-terminus or C-terminus of the chain. |
The model and post-processing
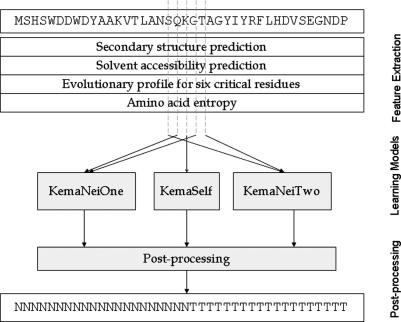
Figure 2 shows the architecture of KemaDom which integrates three binary classification sub-models, KemaSelf, KemaNeiOne and KemaNeiTwo. SVM with probability estimates is used to work out the probability of a residue belonging to boundary residue class, PKemaSelf, PKemaNeiOne and PKemaNeiTwo. The free online tool, libsvm (http://www.csie.ntu.edu.tw/cjlin/libsvmtools/ http://www.csie.ntu.edu.tw/~cjlin/libsvmtools/), is modified for domain prediction purpose. Among the classical kernels, the radial basic function (RBF) is adopted because of its superior performance in generalization ability and convergence speed (18). After the kernel selection, the parameters C and γ are determined as C = 4 and γ = 2, separately.
Figure 2.
The architecture of KemaDom for domain prediction.
A residue can be assigned into boundary residue class with the probability P = max{PKemaSelf, PKemaNeiOne, PKemaNeiTwo} and non-boundary residue class with 1 − P. As we know, the output of the learning model is quite noisy. So we smooth the result by averaging the probabilities of three consecutive residues. To reduce the influence of false signals, we regard that any two boundary residues with distance d ≤ 10 belong to the same domain boundary region. This assumption is reasonable because the reliable domain boundaries can be accepted within 20 residues of the true domain boundary annotated in the CATH database (4,9–11). In addition, boundary residues with no neighboring boundary residues or with the distance <10 from the start position of a chain are ignored while computing the number of domains.
RESULTS AND DISCUSSION
In this section, we test our model and compare its performance with other methods. The measurements of sensitivity (SN) and specificity (SP) are the same with the classical one used in CASP 6 and CAFASP 4. The overall accuracy Acc is the number of correctly predicted chains over the total number of chains. Eightfold cross validation is used to measure the performance.
To provide a baseline to compare the result of KemaDom, we run the random control prediction algorithm as in Ref. (9) on the same dataset. First, the dataset is randomly divided into eight subsets. Then, the number of domains for proteins in each subset are predicted according to the composition of domain numbers in remaining subsets. We repeat this test 100 times and average over the results.
Performance of KemaDom and its sub-models
The results are shown in Table 2, where 1D denotes single-domain chains and 2D denotes two-domain chains. KemaSelf achieves 3% higher Acc, 13% (14%) higher 2D SN and 11% (13%) higher 2D SP than KemaNeiOne (KemaNeiTwo). KemaNeiOne has 1% higher 2D SN and 2% higher 2D SP than KemaNeiTwo. This implies that the 1-neighboring residue information contribute more to identifying boundary residues than the 2-neighboring residue information does. After combining these three sub-models, KemaDom improves Acc up to 76%. And the sensitivity and specificity for single-domain chains are 88 and 83%, respectively. Those for two-domain chains increase to 41 and 57%, separately. In contrary, random control prediction method correctly predicts only 74% single-domain chains and 26% two-domain chains. These results show that the neighboring residue information can be used to improve the domain prediction and KemaDom is more effective than random control prediction method.
Table 2.
Performance of KemaDom and sub-models
| Model/Sub-model | 1D SN | 1D SP | 2D SN | 2D SP | Acc |
|---|---|---|---|---|---|
| KemaDom | 0.88 | 0.83 | 0.41 | 0.57 | 0.76 |
| KemaSelf | 0.89 | 0.81 | 0.36 | 0.55 | 0.74 |
| KemaNeiOne | 0.90 | 0.79 | 0.23 | 0.44 | 0.71 |
| KemaNeiTwo | 0.90 | 0.79 | 0.22 | 0.42 | 0.71 |
| Baseline | 0.74 | 0.72 | 0.26 | 0.23 | 0.60 |
We also use an individual SVM with a combined feature map of three sub-models to predict domains. The results show that this strategy fails in prediction because only two two-domain chains are correctly predicted and others are all inferred to be single-domain chains. Although no well-established theory of this ensemble technique with different features has been given, the subspace ensemble for supervised learning has been successfully applied in bioinfomatics with a satisfactory result (24).
While predicting domain boundary position, KemaDom only correctly predicts 15% of the two-domain chains and 12% of the multi-domain chains; they are both lower than those of DOMpro, 25 and 20%, respectively. It should be pointed out that the reliable domain boundaries are acceptable within 20 residues of the true domain boundary annotated in CATH and predicting domain boundary locations is more difficult than predicting domain numbers.
Objectively, in order to evaluate the performance of KemaDom, we also test KemaDom against CAFASP 4 dataset, in which there are 41 single-domain chains and 17 two-domain chains. In these chains, KemaDom shows 95% 1D SN, 77% 1D SP, 24% 2D SN and 57% 2D SP. The Acc is 74% and the average overlap score of the two-domain chains is 64.18.
Performance comparison with other predictors
The performance of available ab initio systems can be taken from the previous publications and the website of CAFASP 4 (Table 3). It is easy to see that predicting two-domain or multi-domain chains is more difficult than predicting single-domain chains. The 2D SN varies from 12% (Mateo) to 59% (DOMpro), and the 2D SP ranges from 15% (Mateo) to 60% (Globplot) while the Acc lies between 17% (Biozon) and 76% (KemaDom). Moreover, the selection of training and testing datasets influences the performance of the predictors significantly.
Table 3.
Performance of ab initio predictorsa
| Predictor name | 1D SN | 1D SP | 2D SN | 2D SP | Acc | Dataset |
|---|---|---|---|---|---|---|
| KemaDom | 0.88 | 0.83 | 0.41 | 0.57 | 0.76 | (10) |
| DOMpro | 0.76 | 0.85 | 0.59 | 0.38 | 0.69 | (10) |
| CHOPnetb | 0.42–0.73 | N/A | 0.40–0.59 | N/A | 0.69 | (9) |
| KemaDom | 0.95 | 0.77 | 0.24 | 0.57 | 0.74 | CAFASP 4 |
| DOMpro | 0.85 | 0.76 | 0.35 | 0.50 | 0.70 | CAFASP 4 |
| Biozon | 0.10 | 10.00 | 0.35 | 0.19 | 0.17 | CAFASP 4 |
| Globplot | 0.83 | 0.71 | 0.18 | 0.60 | 0.64 | CAFASP 4 |
| Dompred-DPS | 0.68 | 0.78 | 0.47 | 0.50 | 0.62 | CAFASP 4 |
| Mateo | 0.51 | 0.78 | 0.12 | 0.15 | 0.40 | CAFASP 4 |
aThe values taken from the previous publications and the website of CAFASP 4.
bThe performance of CHOPnet is tested against multiple datasets with cross validation of networks; SP values are not shown in their paper and are denoted by N/A in this table.
Compared with DOMpro, KemaDom achieves 19% higher 2D SP and 7% higher Acc on the CATH dataset though it has 18% lower 2D SN. Similarly, on CAFASP 4 dataset, KemaDom has 11% lower 2D SN but 7% higher 2D SP than DOMpro. Obviously, KemaDom achieves a good Acc because of its high 1D SN. On this point, we can not conclude that our method is better or worse than the other methods because the knowledge is still not sufficient for discriminating the boundary residues exactly.
WEB SERVER: KemaDom
The web server can be accessed from http://www.iipl.fudan.edu.cn/lschen/kemadom.htm. and http://www.iipl.fudan.edu.cn/~lschen/kemadom.htm. This system is mainly composed of two subsystems, the background system and the interface system.
The background system is implemented by Perl including package BioPerl and CGI script. The whole processing flowchart of this system can be summarized as the following steps: (i) a remote user submits a target sequence to the server; (ii) a PSSM profile for the sequence is generated by PSI-blast (25) against the non-redundant (nr) database; and (iii) secondary structure prediction and solvent accessibility prediction are performed by SSpro (26) and ACCpro (27), respectively; (iv) a Perl script generates the feature vectors for all the residues of the input sequence; (v) boundary residues prediction is executed with the feature vectors against the trained model. (vi) post-processing is done for the raw output; and (vii) KemaDom sends the result to the user.
The interface system is written with HTML language. KemaDom provides a friendly interface (Figure 3). Users should submit sequences with the format which BioPerl (Bio::SeqIO) can recognize. Also, the email address and the customized job name are required in submission. The only constraint is that protein sequence to be predicted should contain >30 residues.
Figure 3.
The interface of KemaDom. The Email address and the customized job name are required. The target sequence should be input with format which BioPerl (Bio::SeqIO) can recognize.
CONCLUSION
In this paper, we have presented a novel domain prediction server, KemaDom, modeling the local context information. As a domain prediction server, it is powerful and easy to use. This method is a good option for domain prediction compared with the existing methods.
Acknowledgments
The authors would like to thank both anonymous reviewers for their constructive comments and Yanqiu Chen for improving the presentation of the manuscript. The authors also would like to thank Yu Xue from USTC, People Republic of China for his valuable discussion. This work is supported by the Major Research Program of the National Natural Science Foundation of China (No. 60496324) and the National Natural Science Foundation of China (No. 60303009). The authors would also like to acknowledge the grant of the Open Program of Beijing Municipal Key laboratory (No. KP701200372). Funding to pay the Open Access publication charges for this article was provided by Shanghai Key Laboratory of Intelligent Information Processing, Fudan University.
Conflict of interest statement. None declared.
REFERENCES
- 1.Vogel C., Teichmann S.A., Pereira-Leal J. The relationship between domain duplication and recombination. J. Mol. Biol. 2004;346:355–365. doi: 10.1016/j.jmb.2004.11.050. [DOI] [PubMed] [Google Scholar]
- 2.Chivian D., Kim D.E., Malmstrom L., Bradley P., Robertson T., Murphy P., Strauss C.E., Bon-neau R., Rohl C.A., Baker D. Automated prediction of CASP-5 structures using the Robetta server. Proteins. 2003;53:524–533. doi: 10.1002/prot.10529. [DOI] [PubMed] [Google Scholar]
- 3.Heger A., Holm L. Exhaustive enumeration of protein domain families. J. Mol. Biol. 2003;328:749–776. doi: 10.1016/s0022-2836(03)00269-9. [DOI] [PubMed] [Google Scholar]
- 4.Marsden R.L., McGuffin L.J., Jones D.T. Rapid protein domain assignment from amino acid sequence using predicted secondary structure. Protein Sci. 2002;11:2814–2824. doi: 10.1110/ps.0209902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.von Ohsen N., Sommer I., Zimmer R., Lengauer T. Arby: automatic protein structure prediction using profile–profile alignment and confidence measures. Bioinformatics. 2004;20:2228–2235. doi: 10.1093/bioinformatics/bth232. [DOI] [PubMed] [Google Scholar]
- 6.Zdobnov E.M., Apweiler R. InterProScan: an integration platform for the signature-recognition methods in InterPro. Bioinformatics. 2001;17:847–848. doi: 10.1093/bioinformatics/17.9.847. [DOI] [PubMed] [Google Scholar]
- 7.Gewehr J.E., Zimmer R. SSEP-Domain: protein domain prediction by alignment of secondary structure elements and profiles. Bioinformatics. 2006;22:181–187. doi: 10.1093/bioinformatics/bti751. [DOI] [PubMed] [Google Scholar]
- 8.Nagarajan N., Yona G. Automatic prediction of protein domains from sequence information using a hybrid learning system. Bioinformatics. 2004;20:1335–1360. doi: 10.1093/bioinformatics/bth086. [DOI] [PubMed] [Google Scholar]
- 9.Liu J., Rost B. Sequence-based prediction of protein domains. Nucleic Acids Res. 2004;32:3522–3530. doi: 10.1093/nar/gkh684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cheng J., Sweredoski M.J., Baldi P. Data Mining and Knowledge Discovery. 2005. DOMpro: protein domain prediction using profiles,secondary structure, relative solvent accessibility, and recursive neural networks. in press. [Google Scholar]
- 11.Bryson K., McGuffin L.J., Marsden R.L., Ward J.J., Sodhi J.S., Jones D.T. Protein structure prediction servers at University College London. Nucleic Acids Res. 2005;33:W36–W38. doi: 10.1093/nar/gki410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Linding R., Russell R.B., Neduva V., Gibson T.J. GlobPlot: exploring protein sequences for globularity and disorder. Nucleic Acids Res. 2003;31:3701–3708. doi: 10.1093/nar/gkg519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lexa M., Valle G. PRIMEX: rapid identification of oligonucleotide matches in whole genomes. Bioinformatics. 2003;19:2486–2488. doi: 10.1093/bioinformatics/btg350. [DOI] [PubMed] [Google Scholar]
- 14.Saini H.K., Fischer D. Meta-DP: domain prediction meta server. Bioinformatics. 2005;21:2917–2920. doi: 10.1093/bioinformatics/bti445. [DOI] [PubMed] [Google Scholar]
- 15.Orengo C.A., Bray J.E., Buchan D.W., Harrison A., Lee D., Perl F.M., Sillitoe I., Todd A.E., Thornton J.M. The CATH protein family database: a resource for structural and functional annotation of genomes. Proteomics. 2002;2:11–21. [PubMed] [Google Scholar]
- 16.Murzin A.G., Brenner S.E., Hubbard T., Chothia C. SCOP: a structural classification of proteins database for the investigation of sequences and structures. J. Mol. Biol. 1995;247:536–540. doi: 10.1006/jmbi.1995.0159. [DOI] [PubMed] [Google Scholar]
- 17.Holm L., Sander C. Touring protein fold space with Dali/FSSP. Nucleic Acids Res. 1998;26:316–319. doi: 10.1093/nar/26.1.316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vapnik V. Statistical Learning Theory. NY: John Wiley and Sons. Ins.; 1998. [Google Scholar]
- 19.Busuttil S., Abela J., Pace G.J. Support vector machines with profile-based kernels for remote protein homology detection. Genome Inform. Ser. Workshop Genome Inform. 2004;15:191–200. [PubMed] [Google Scholar]
- 20.Saigo H., Vert J.P., Ueda N., Akutsu T. Protein homology detection using string alignment kernels. Bioinformatics. 2004;20:1682–1689. doi: 10.1093/bioinformatics/bth141. [DOI] [PubMed] [Google Scholar]
- 21.Hua S., Sun Z. A novel method of protein secondary structure prediction with high segment overlap measure: support vector machine approach. J. Mol. Biol. 2001;308:397–407. doi: 10.1006/jmbi.2001.4580. [DOI] [PubMed] [Google Scholar]
- 22.Guo J., Chen H., Sun Z., Lin Y. a novel method for protein secondary structure prediction using dual-layer SVM and profiles. Proteins. 2004;54:738–743. doi: 10.1002/prot.10634. [DOI] [PubMed] [Google Scholar]
- 23.Ferran E.A., Pflugfelder B., Ferrara P. Self-organized neural maps of human protein sequences. Protein Sci. 1994;3:507–521. doi: 10.1002/pro.5560030316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bertoni A., Folgieri R., Valentini G. Bio-molecular cancer prediction with random subspace ensembles of Support Vector Machines. Neurocomputing. 2005;63C:535–539. [Google Scholar]
- 25.Altschul S.F., Madden T.L., Schaffer A.A., Zhang J., Zhang Z., Miller W., Lipman D.J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Baldi P., Pollastri G. The principled design of large-scale recursive neural network architectures-DAG-RNNs and the protein structure prediction problem. J. Mach. Learn. Res. 2003;4:575–602. [Google Scholar]
- 27.Pollastri G., Baldi P., Fariselli P., Casadio R. Prediction of coordination number and relative solvent accessibility in proteins. Proteins. 2002;47:142–153. doi: 10.1002/prot.10069. [DOI] [PubMed] [Google Scholar]